Catalogue of Cancer Genes
The interactome of Cancer Genes
About Catalogue of Cancer GenesIdentifying how cancer driver mutations affect the network properties of the human molecular interactome will help researchers to better understand genotype-to-phenotype relationships in tumorigenesis. In this work, we utilized the large scale human protein interactome network and systematically studied the network topology, somatic mutation, evolutionary rate, and evolutionary origin of cancer genes involved in tumorigenesis. Specially, we constructed a Catalogue of Cancer Genes (CCG) resource, including 4,050 cancer genes with functional and network annotations. We constructed five different yet complementary protein interactomes: a physical Protein-Protein Interaction Network (PPIN) with 113,473 physical PPI pairs connecting 13,579 proteins, an atomic resolution Three-Dimensional structural Protein-Protein Interaction Network (3DPPIN) with 4,278 3DPPI pairs connecting 2,609 proteins, a Kinase-Substrate Interaction Network (KSIN) with 7,543 KSI pairs connecting 379 kinases and 1,961 non-kinase substrate proteins, an INnate immunity related Protein-Protein Interaction Network (INPPIN) with 6,009 INPPI pairs connecting 2,787 proteins, and a massive computational predicted Protein-Protein Interaction Network (CPPIN) with 474,017 unique PPI pairs connecting 16,166 proteins. A total of 3,650 CGA proteins (90.1%) were successfully mapped with at least one interaction of any type, and 10,773 of these interacting partners were recruited. Overall, the final cancer protein interactome includes 95,578 unique interactions, among which there are 79,755 physical PPI pairs, 3,487 3DPPI pairs, 7,226 KSI pairs, and 5,110 INPPI pairs (Fig. 1).
|
||
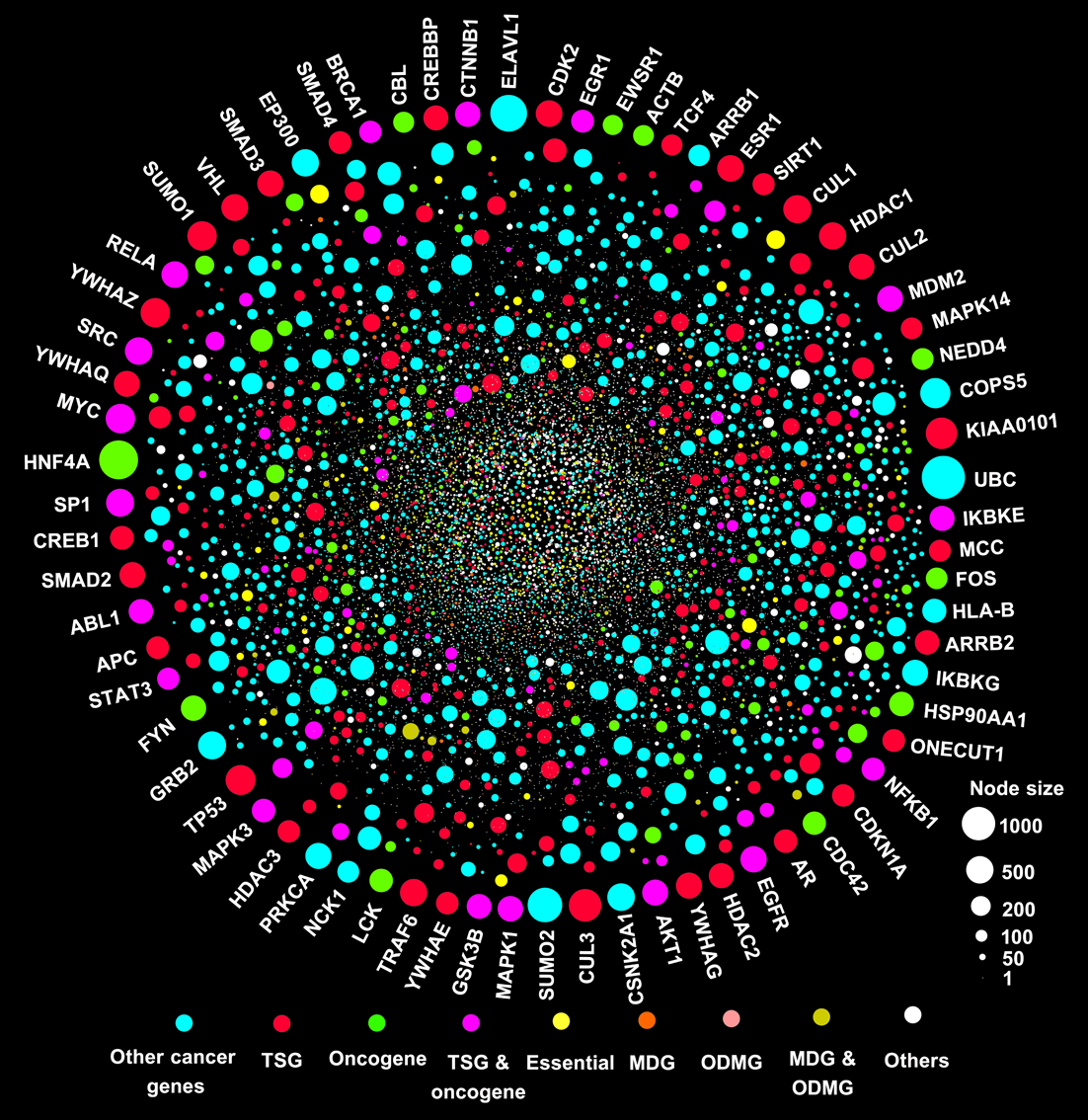
|
||
|
We found distinct network and evolutionary signatures of cancer proteins. Cancer proteins reflected the unique network centrality, which tends to be comprised of central hubs rather than peripheral nodes, regardless of whether they are essential or non-essential proteins. Furthermore, cancer proteins indicate a stronger purifying selection, a tendency to be ancient, and, specifically, come into existence with early metazoans, yet they are younger than Mendelian and orphan disease proteins. Protein evolutionary origin was identified as a major determinant of network topology. We found a positive correlation between protein connectivity and the number of somatic mutations from The Cancer Genome Atlas and the Catalogue of Somatic Mutations in Cancer database. Our study revealed that the somatic mutational network-attacking perturbations to central hubs of the cancer interactome are a major feature of tumor emergence and evolution. This finding elucidates rapid tumorigenesis initiated by a few driver mutations.
|
