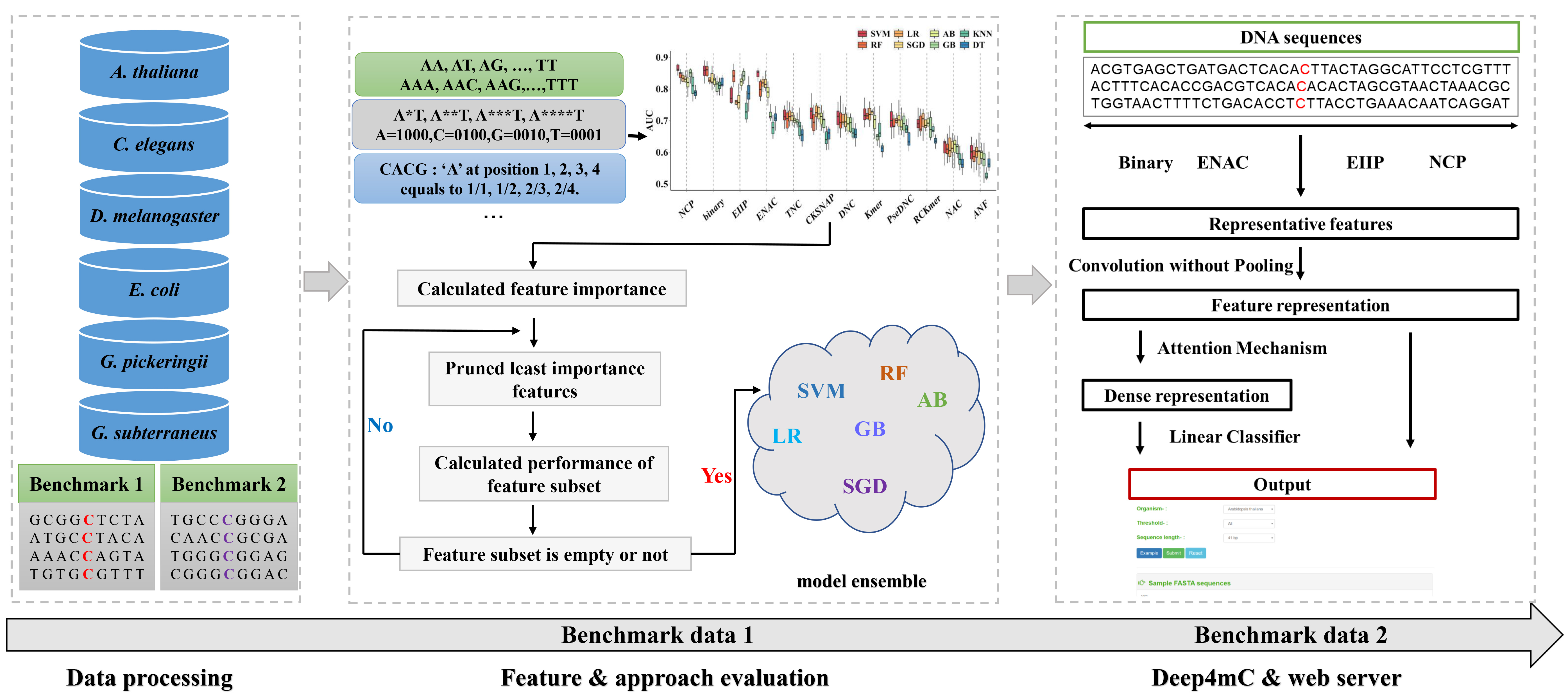
Introduction of the Deep4mC :
DNA N4-methylcytosine (4mC) represents a novel epigenetic modification in genomes of diverse species. By adding a methyl group to the 4th position of cytosine in a DNA sequence, 4mC modification not only plays an important role in the regulation of DNA replication, cell cycle and gene expression, but also participates in genome stabilization, recombination and evolution. The identification of new 4mC loci is fundamental for our understanding of the molecular mechanisms and regulatory roles of 4mC. Currently, the experimental approaches for 4mC identification are very labor-intensive and expensive; therefore, computational prediction of 4mC sites may serve as a rapid and much less expensive approach.
In this study, we first reviewed the current progress in the computational prediction of 4mC sites and systematically evaluated the predictive capacity of eight conventional machine-learning algorithms as well as 12 feature types common used in previous studies in six species. Using a representative benchmark data set, we also investigate the contribution of feature selection and stacking approach to the model construction, indicating feature optimization and proper reinforcement learning could improve the performance. We next re-collected 285,851 experimentally identified 4mC sites in the six species genomes and developed a novel deep learning-based 4mC site predictor, namely Deep4mC, with convolutional neural networks (CNNs) using four representative features. For species with small number of samples, we extended our deep learning framework with a bootstrapping method. Our evaluation indicated that Deep4mC could obtain high accuracy and robust performance with the average Area Under Curve (AUC) values more than 0.9 of multiple cross-validations across different species (from 0.9005 to 0.9722). By comparison, Deep4mC achieved great AUC values improvement from 10.14% to 46.21% compared to previous tools in the independent data set in different species.