Gene Page: HDAC5
Summary ?
| GeneID | 10014 |
| Symbol | HDAC5 |
| Synonyms | HD5|NY-CO-9 |
| Description | histone deacetylase 5 |
| Reference | MIM:605315|HGNC:HGNC:14068|Ensembl:ENSG00000108840|HPRD:09246|Vega:OTTHUMG00000181806 |
| Gene type | protein-coding |
| Map location | 17q21 |
| Pascal p-value | 0.363 |
| Sherlock p-value | 0.975 |
| Fetal beta | 0.65 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
| Support | CompositeSet Darnell FMRP targets Chromatin Remodeling Genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg14776943 | 17 | 42201167 | HDAC5 | 6.6E-8 | -0.01 | 1.63E-5 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs11984825 | chr8 | 1201727 | HDAC5 | 10014 | 0.17 | trans |
Section II. Transcriptome annotation
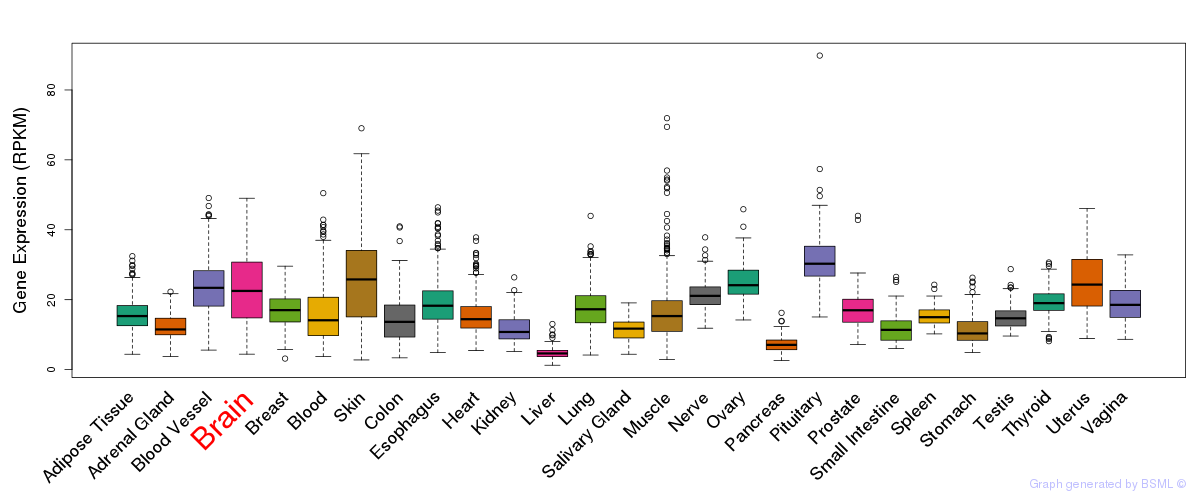
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SUCLA2 | 0.91 | 0.91 |
| UBE2E2 | 0.91 | 0.91 |
| TMEM66 | 0.91 | 0.92 |
| ATL1 | 0.90 | 0.91 |
| RTN3 | 0.90 | 0.91 |
| GHITM | 0.90 | 0.91 |
| REEP5 | 0.90 | 0.91 |
| MOAP1 | 0.90 | 0.92 |
| NAPG | 0.90 | 0.92 |
| COQ10B | 0.90 | 0.91 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.18 | -0.51 | -0.30 |
| AF347015.21 | -0.48 | -0.21 |
| AC010300.1 | -0.46 | -0.44 |
| AC098691.2 | -0.44 | -0.32 |
| FAM159B | -0.44 | -0.49 |
| AL022328.1 | -0.44 | -0.41 |
| AC135724.1 | -0.43 | -0.47 |
| AC100783.1 | -0.43 | -0.36 |
| NSBP1 | -0.43 | -0.37 |
| C10orf108 | -0.42 | -0.37 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ANKRD11 | ANCO-1 | LZ16 | T13 | ankyrin repeat domain 11 | - | HPRD | 15184363 |
| BCL6 | BCL5 | BCL6A | LAZ3 | ZBTB27 | ZNF51 | B-cell CLL/lymphoma 6 | - | HPRD,BioGRID | 11929873 |
| BCOR | ANOP2 | FLJ20285 | FLJ38041 | KIAA1575 | MAA2 | MCOPS2 | MGC131961 | MGC71031 | BCL6 co-repressor | - | HPRD | 10898795 |
| CBX5 | HP1 | HP1A | chromobox homolog 5 (HP1 alpha homolog, Drosophila) | - | HPRD,BioGRID | 12242305 |
| CTBP1 | BARS | MGC104684 | C-terminal binding protein 1 | Reconstituted Complex | BioGRID | 11022042 |
| EEF1G | EF1G | GIG35 | eukaryotic translation elongation factor 1 gamma | Two-hybrid | BioGRID | 16169070 |
| GABARAP | FLJ25768 | MGC120154 | MGC120155 | MM46 | GABA(A) receptor-associated protein | Two-hybrid | BioGRID | 16169070 |
| GATA1 | ERYF1 | GF-1 | GF1 | NFE1 | GATA binding protein 1 (globin transcription factor 1) | - | HPRD,BioGRID | 14668799 |
| GATA2 | MGC2306 | NFE1B | GATA binding protein 2 | - | HPRD,BioGRID | 11567998 |
| GPS2 | AMF-1 | MGC104294 | MGC119287 | MGC119288 | MGC119289 | G protein pathway suppressor 2 | Affinity Capture-Western | BioGRID | 11931768 |
| HDAC3 | HD3 | RPD3 | RPD3-2 | histone deacetylase 3 | - | HPRD,BioGRID | 11804585 |
| HDAC3 | HD3 | RPD3 | RPD3-2 | histone deacetylase 3 | - | HPRD | 11466315 |
| HR | ALUNC | AU | HSA277165 | hairless homolog (mouse) | Affinity Capture-Western | BioGRID | 11641275 |
| IKZF1 | Hs.54452 | IK1 | IKAROS | LYF1 | PRO0758 | ZNFN1A1 | hIk-1 | IKAROS family zinc finger 1 (Ikaros) | Affinity Capture-Western | BioGRID | 12015313 |
| IKZF2 | HELIOS | MGC34330 | ZNF1A2 | ZNFN1A2 | IKAROS family zinc finger 2 (Helios) | Affinity Capture-Western | BioGRID | 12015313 |
| IKZF3 | AIO | AIOLOS | ZNFN1A3 | IKAROS family zinc finger 3 (Aiolos) | Affinity Capture-Western | BioGRID | 12015313 |
| IKZF4 | EOS | KIAA1782 | ZNFN1A4 | IKAROS family zinc finger 4 (Eos) | Affinity Capture-Western | BioGRID | 12015313 |
| MAP3K7IP2 | FLJ21885 | KIAA0733 | TAB2 | mitogen-activated protein kinase kinase kinase 7 interacting protein 2 | Reconstituted Complex | BioGRID | 12150997 |
| MEF2A | ADCAD1 | RSRFC4 | RSRFC9 | myocyte enhancer factor 2A | - | HPRD,BioGRID | 10748098 |
| MEF2D | DKFZp686I1536 | myocyte enhancer factor 2D | Affinity Capture-Western | BioGRID | 12896970 |
| NCOR1 | KIAA1047 | MGC104216 | N-CoR | TRAC1 | hCIT529I10 | hN-CoR | nuclear receptor co-repressor 1 | - | HPRD,BioGRID | 10640275 |
| NCOR2 | CTG26 | SMRT | SMRTE | SMRTE-tau | TNRC14 | TRAC-1 | TRAC1 | nuclear receptor co-repressor 2 | - | HPRD | 10869435 |
| NCOR2 | CTG26 | SMRT | SMRTE | SMRTE-tau | TNRC14 | TRAC-1 | TRAC1 | nuclear receptor co-repressor 2 | Reconstituted Complex Two-hybrid | BioGRID | 10640275 |
| NRIP1 | FLJ77253 | RIP140 | nuclear receptor interacting protein 1 | Co-localization Reconstituted Complex | BioGRID | 15060175 |
| PRKD1 | PKC-MU | PKCM | PKD | PRKCM | protein kinase D1 | Affinity Capture-Western | BioGRID | 15367659 |
| RUNX3 | AML2 | CBFA3 | FLJ34510 | MGC16070 | PEBP2aC | runt-related transcription factor 3 | - | HPRD | 15138260 |
| RUVBL2 | CGI-46 | ECP51 | INO80J | REPTIN | RVB2 | TIH2 | TIP48 | TIP49B | RuvB-like 2 (E. coli) | RUVBL2 (reptin) interacts with an unspecified isoform of HDAC5. | BIND | 15829968 |
| SFN | YWHAS | stratifin | - | HPRD | 15367659 |
| SUV39H1 | KMT1A | MG44 | SUV39H | suppressor of variegation 3-9 homolog 1 (Drosophila) | - | HPRD,BioGRID | 12242305 |
| TBL1X | EBI | SMAP55 | TBL1 | transducin (beta)-like 1X-linked | Affinity Capture-Western | BioGRID | 11931768 |
| TNFSF10 | APO2L | Apo-2L | CD253 | TL2 | TRAIL | tumor necrosis factor (ligand) superfamily, member 10 | An unspecifed isoform of HDAC5 interacts with the TNFSF10 promoter. | BIND | 15619633 |
| YWHAB | GW128 | HS1 | KCIP-1 | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, beta polypeptide | - | HPRD,BioGRID | 10869435 |
| YWHAE | 14-3-3E | FLJ45465 | KCIP-1 | MDCR | MDS | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, epsilon polypeptide | - | HPRD,BioGRID | 10869435 |
| YWHAQ | 14-3-3 | 1C5 | HS1 | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide | Affinity Capture-Western Two-hybrid | BioGRID | 15367659 |
| ZBTB16 | PLZF | ZNF145 | zinc finger and BTB domain containing 16 | - | HPRD,BioGRID | 11929873 |15467736 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| BIOCARTA CARM ER PATHWAY | 35 | 27 | All SZGR 2.0 genes in this pathway |
| BIOCARTA HDAC PATHWAY | 32 | 25 | All SZGR 2.0 genes in this pathway |
| BIOCARTA ETS PATHWAY | 18 | 12 | All SZGR 2.0 genes in this pathway |
| BIOCARTA PGC1A PATHWAY | 26 | 20 | All SZGR 2.0 genes in this pathway |
| PID HDAC CLASSII PATHWAY | 34 | 27 | All SZGR 2.0 genes in this pathway |
| PID HDAC CLASSI PATHWAY | 66 | 50 | All SZGR 2.0 genes in this pathway |
| REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | 46 | 28 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY NOTCH1 | 70 | 46 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY NOTCH | 103 | 64 | All SZGR 2.0 genes in this pathway |
| DAVICIONI PAX FOXO1 SIGNATURE IN ARMS UP | 59 | 38 | All SZGR 2.0 genes in this pathway |
| ZHONG RESPONSE TO AZACITIDINE AND TSA UP | 183 | 119 | All SZGR 2.0 genes in this pathway |
| DAVICIONI MOLECULAR ARMS VS ERMS UP | 332 | 228 | All SZGR 2.0 genes in this pathway |
| DAVICIONI TARGETS OF PAX FOXO1 FUSIONS UP | 255 | 177 | All SZGR 2.0 genes in this pathway |
| DAVICIONI RHABDOMYOSARCOMA PAX FOXO1 FUSION UP | 64 | 37 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION DN | 329 | 219 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA UP | 1821 | 933 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE EPIDERMIS DN | 508 | 354 | All SZGR 2.0 genes in this pathway |
| RIZ ERYTHROID DIFFERENTIATION 12HR | 43 | 35 | All SZGR 2.0 genes in this pathway |
| PEREZ TP53 TARGETS | 1174 | 695 | All SZGR 2.0 genes in this pathway |
| BUYTAERT PHOTODYNAMIC THERAPY STRESS UP | 811 | 508 | All SZGR 2.0 genes in this pathway |
| LOPEZ MBD TARGETS | 957 | 597 | All SZGR 2.0 genes in this pathway |
| SAKAI TUMOR INFILTRATING MONOCYTES UP | 27 | 19 | All SZGR 2.0 genes in this pathway |
| PENG LEUCINE DEPRIVATION UP | 142 | 93 | All SZGR 2.0 genes in this pathway |
| PEART HDAC PROLIFERATION CLUSTER UP | 57 | 35 | All SZGR 2.0 genes in this pathway |
| PENG RAPAMYCIN RESPONSE UP | 203 | 130 | All SZGR 2.0 genes in this pathway |
| THEILGAARD NEUTROPHIL AT SKIN WOUND DN | 225 | 163 | All SZGR 2.0 genes in this pathway |
| CHANG CORE SERUM RESPONSE DN | 209 | 137 | All SZGR 2.0 genes in this pathway |
| HOFFMANN PRE BI TO LARGE PRE BII LYMPHOCYTE UP | 36 | 24 | All SZGR 2.0 genes in this pathway |
| SHEDDEN LUNG CANCER GOOD SURVIVAL A4 | 196 | 124 | All SZGR 2.0 genes in this pathway |
| HAN SATB1 TARGETS UP | 395 | 249 | All SZGR 2.0 genes in this pathway |
| SHAFFER IRF4 TARGETS IN MYELOMA VS MATURE B LYMPHOCYTE | 101 | 76 | All SZGR 2.0 genes in this pathway |
| QI HYPOXIA | 140 | 96 | All SZGR 2.0 genes in this pathway |
| KASLER HDAC7 TARGETS 1 UP | 194 | 133 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS UP | 745 | 475 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION DN | 1080 | 713 | All SZGR 2.0 genes in this pathway |