Gene Page: GNE
Summary ?
| GeneID | 10020 |
| Symbol | GNE |
| Synonyms | DMRV|GLCNE|IBM2|NM|Uae1 |
| Description | glucosamine (UDP-N-acetyl)-2-epimerase/N-acetylmannosamine kinase |
| Reference | MIM:603824|HGNC:HGNC:23657|Ensembl:ENSG00000159921|HPRD:04825|Vega:OTTHUMG00000019899 |
| Gene type | protein-coding |
| Map location | 9p13.3 |
| Pascal p-value | 0.012 |
| Sherlock p-value | 0.382 |
| DEG p-value | DEG:Sanders_2014:DS1_p=-0.143:DS1_beta=0.048700:DS2_p=5.77e-01:DS2_beta=0.028:DS2_FDR=7.80e-01 |
| Fetal beta | 1.034 |
| DMG | 1 (# studies) |
| eGene | Putamen basal ganglia |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DEG:Sanders_2013 | Microarray | Whole-genome gene expression profiles using microarrays on lymphoblastoid cell lines (LCLs) from 413 cases and 446 controls. | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg21177599 | 9 | 36258839 | GNE | 6.34E-8 | -0.006 | 1.57E-5 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs10117275 | 9 | 36265159 | GNE | ENSG00000159921.10 | 5.3515E-6 | 0.04 | 11894 | gtex_brain_putamen_basal |
| rs7863247 | 9 | 36273828 | GNE | ENSG00000159921.10 | 5.34717E-6 | 0.04 | 3225 | gtex_brain_putamen_basal |
| rs7875447 | 9 | 36276857 | GNE | ENSG00000159921.10 | 5.26712E-6 | 0.04 | 196 | gtex_brain_putamen_basal |
| rs2182817 | 9 | 36280788 | GNE | ENSG00000159921.10 | 5.16938E-6 | 0.04 | -3735 | gtex_brain_putamen_basal |
| rs4879975 | 9 | 36286706 | GNE | ENSG00000159921.10 | 4.99855E-6 | 0.04 | -9653 | gtex_brain_putamen_basal |
| rs7045725 | 9 | 36293918 | GNE | ENSG00000159921.10 | 4.19781E-6 | 0.04 | -16865 | gtex_brain_putamen_basal |
Section II. Transcriptome annotation
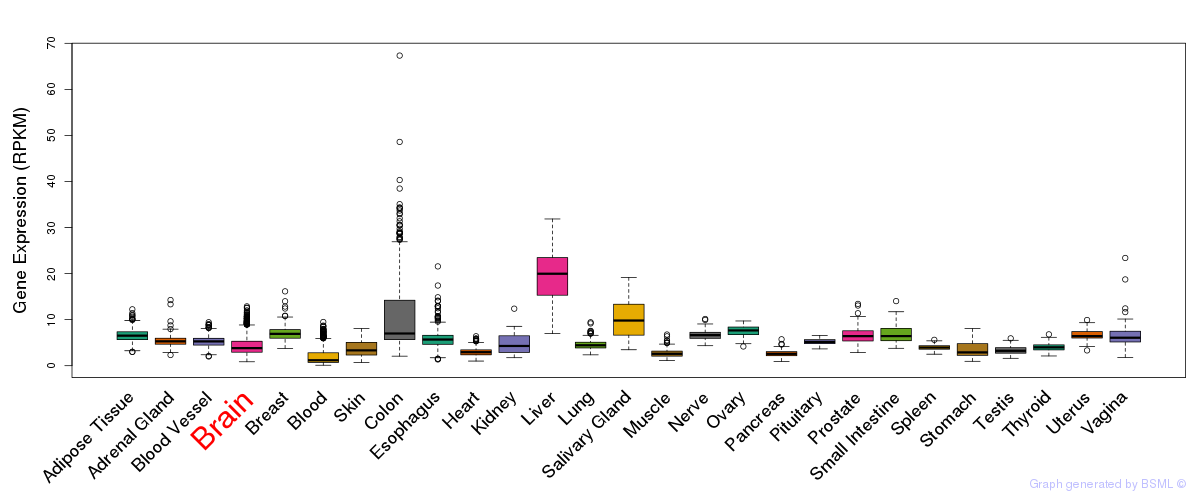
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| TMEM87A | 0.74 | 0.75 |
| DIS3L | 0.72 | 0.69 |
| TMCO3 | 0.72 | 0.71 |
| ANUBL1 | 0.70 | 0.72 |
| RPS6KC1 | 0.70 | 0.69 |
| GALC | 0.70 | 0.70 |
| ANKIB1 | 0.70 | 0.73 |
| ZNF621 | 0.69 | 0.73 |
| ACSL1 | 0.69 | 0.76 |
| PIGM | 0.68 | 0.72 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.21 | -0.53 | -0.53 |
| CXCL14 | -0.50 | -0.50 |
| MT-CO2 | -0.49 | -0.50 |
| AF347015.31 | -0.48 | -0.49 |
| IFI27 | -0.47 | -0.46 |
| AF347015.8 | -0.45 | -0.46 |
| CLEC2B | -0.45 | -0.44 |
| AP000679.1 | -0.44 | -0.47 |
| HIGD1B | -0.44 | -0.44 |
| MT-CYB | -0.44 | -0.44 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG AMINO SUGAR AND NUCLEOTIDE SUGAR METABOLISM | 44 | 30 | All SZGR 2.0 genes in this pathway |
| DAVICIONI MOLECULAR ARMS VS ERMS UP | 332 | 228 | All SZGR 2.0 genes in this pathway |
| GARGALOVIC RESPONSE TO OXIDIZED PHOSPHOLIPIDS GREEN UP | 23 | 19 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC3 TARGETS DN | 536 | 332 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA UP | 1821 | 933 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE KERATINOCYTE DN | 485 | 334 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER SERUM RESPONSE DN | 712 | 443 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 DN | 855 | 609 | All SZGR 2.0 genes in this pathway |
| UEDA PERIFERAL CLOCK | 169 | 111 | All SZGR 2.0 genes in this pathway |
| FRASOR RESPONSE TO ESTRADIOL DN | 82 | 52 | All SZGR 2.0 genes in this pathway |
| GENTILE UV RESPONSE CLUSTER D4 | 55 | 37 | All SZGR 2.0 genes in this pathway |
| LU AGING BRAIN UP | 262 | 186 | All SZGR 2.0 genes in this pathway |
| GENTILE UV HIGH DOSE DN | 312 | 203 | All SZGR 2.0 genes in this pathway |
| DAZARD RESPONSE TO UV NHEK DN | 318 | 220 | All SZGR 2.0 genes in this pathway |
| BAELDE DIABETIC NEPHROPATHY DN | 434 | 302 | All SZGR 2.0 genes in this pathway |
| DAZARD UV RESPONSE CLUSTER G6 | 153 | 112 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR UP | 953 | 554 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR UP | 783 | 442 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 3 | 720 | 440 | All SZGR 2.0 genes in this pathway |
| BONOME OVARIAN CANCER SURVIVAL SUBOPTIMAL DEBULKING | 510 | 309 | All SZGR 2.0 genes in this pathway |
| QI PLASMACYTOMA DN | 100 | 63 | All SZGR 2.0 genes in this pathway |
| CROMER TUMORIGENESIS DN | 51 | 29 | All SZGR 2.0 genes in this pathway |
| SWEET LUNG CANCER KRAS UP | 491 | 316 | All SZGR 2.0 genes in this pathway |
| CHEN METABOLIC SYNDROM NETWORK | 1210 | 725 | All SZGR 2.0 genes in this pathway |
| CHIANG LIVER CANCER SUBCLASS PROLIFERATION DN | 179 | 97 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH T 8 21 TRANSLOCATION | 368 | 247 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA DN | 267 | 160 | All SZGR 2.0 genes in this pathway |
| GABRIELY MIR21 TARGETS | 289 | 187 | All SZGR 2.0 genes in this pathway |
| GUO TARGETS OF IRS1 AND IRS2 | 98 | 67 | All SZGR 2.0 genes in this pathway |
| ACEVEDO FGFR1 TARGETS IN PROSTATE CANCER MODEL UP | 289 | 184 | All SZGR 2.0 genes in this pathway |
| LIM MAMMARY LUMINAL PROGENITOR UP | 58 | 30 | All SZGR 2.0 genes in this pathway |