Gene Page: CDH10
Summary ?
| GeneID | 1008 |
| Symbol | CDH10 |
| Synonyms | - |
| Description | cadherin 10 |
| Reference | MIM:604555|HGNC:HGNC:1749|Ensembl:ENSG00000040731|HPRD:05187|Vega:OTTHUMG00000090667 |
| Gene type | protein-coding |
| Map location | 5p14.2 |
| Pascal p-value | 0.004 |
| Sherlock p-value | 0.715 |
| Fetal beta | -0.765 |
| Support | CELL ADHESION AND TRANSSYNAPTIC SIGNALING G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL G2Cdb.human_BAYES-COLLINS-MOUSE-PSD-CONSENSUS |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
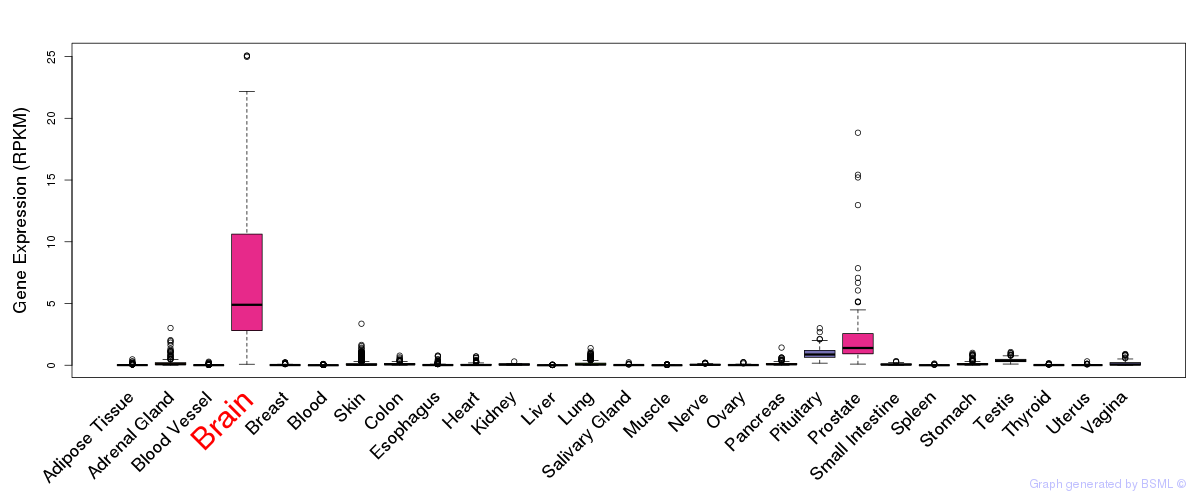
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| GABRA6 | 0.76 | 0.22 |
| CBLN3 | 0.74 | 0.07 |
| PCP2 | 0.54 | 0.27 |
| RP13-102H20.1 | 0.51 | 0.23 |
| FAT2 | 0.48 | 0.35 |
| GRM4 | 0.43 | 0.39 |
| CRTAM | 0.42 | 0.24 |
| CDC42BPG | 0.42 | 0.40 |
| RGS3 | 0.36 | 0.39 |
| DOK7 | 0.35 | 0.32 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| C6orf142 | -0.18 | -0.17 |
| FBXW7 | -0.18 | -0.10 |
| DPP4 | -0.18 | -0.13 |
| AC016910.1 | -0.18 | -0.11 |
| GPR22 | -0.18 | -0.13 |
| GTDC1 | -0.18 | -0.14 |
| SATB1 | -0.17 | -0.10 |
| CIDEA | -0.17 | -0.13 |
| SLC26A4 | -0.17 | -0.17 |
| MCTP1 | -0.16 | -0.11 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| REACTOME CELL CELL COMMUNICATION | 120 | 77 | All SZGR 2.0 genes in this pathway |
| REACTOME ADHERENS JUNCTIONS INTERACTIONS | 27 | 20 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CELL JUNCTION ORGANIZATION | 56 | 31 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL JUNCTION ORGANIZATION | 78 | 43 | All SZGR 2.0 genes in this pathway |
| UDAYAKUMAR MED1 TARGETS DN | 240 | 171 | All SZGR 2.0 genes in this pathway |
| XU HGF TARGETS REPRESSED BY AKT1 DN | 95 | 58 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 48HR DN | 428 | 306 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| ZHENG BOUND BY FOXP3 | 491 | 310 | All SZGR 2.0 genes in this pathway |
| ALONSO METASTASIS EMT DN | 5 | 5 | All SZGR 2.0 genes in this pathway |
| ALONSO METASTASIS DN | 26 | 17 | All SZGR 2.0 genes in this pathway |
| CHANDRAN METASTASIS DN | 306 | 191 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN NPC ICP WITH H3K4ME3 | 445 | 257 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION DN | 1080 | 713 | All SZGR 2.0 genes in this pathway |
| KATSANOU ELAVL1 TARGETS UP | 169 | 105 | All SZGR 2.0 genes in this pathway |