Gene Page: LRPPRC
Summary ?
| GeneID | 10128 |
| Symbol | LRPPRC |
| Synonyms | CLONE-23970|GP130|LRP130|LSFC |
| Description | leucine rich pentatricopeptide repeat containing |
| Reference | MIM:607544|HGNC:HGNC:15714|Ensembl:ENSG00000138095|HPRD:06343|Vega:OTTHUMG00000152782 |
| Gene type | protein-coding |
| Map location | 2p21 |
| Pascal p-value | 0.99 |
| Sherlock p-value | 0.167 |
| Fetal beta | 0.032 |
| DMG | 2 (# studies) |
| eGene | Anterior cingulate cortex BA24 Caudate basal ganglia Cerebellar Hemisphere Cerebellum Cortex Frontal Cortex BA9 Hippocampus Hypothalamus Nucleus accumbens basal ganglia Putamen basal ganglia Meta |
| Support | G2Cdb.humanPSD G2Cdb.humanPSP CompositeSet |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 2 |
| DMG:Nishioka_2013 | Genome-wide DNA methylation analysis | The authors investigated the methylation profiles of DNA in peripheral blood cells from 18 patients with first-episode schizophrenia (FESZ) and from 15 normal controls. | 2 |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0113 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg26538116 | 2 | 44223328 | LRPPRC | -0.02 | 0.85 | DMG:Nishioka_2013 | |
| cg01913188 | 2 | 44223249 | LRPPRC | 5.68E-8 | -0.009 | 1.46E-5 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs6720846 | 2 | 44123073 | LRPPRC | ENSG00000138095.14 | 7.419E-7 | 0.02 | 100071 | gtex_brain_ba24 |
| rs201507883 | 2 | 44123356 | LRPPRC | ENSG00000138095.14 | 6.232E-7 | 0.02 | 99788 | gtex_brain_ba24 |
| rs3839036 | 2 | 44123672 | LRPPRC | ENSG00000138095.14 | 9.063E-7 | 0.02 | 99472 | gtex_brain_ba24 |
| rs4953032 | 2 | 44128912 | LRPPRC | ENSG00000138095.14 | 1.03E-6 | 0.02 | 94232 | gtex_brain_ba24 |
| rs397870554 | 2 | 44135775 | LRPPRC | ENSG00000138095.14 | 2.164E-6 | 0.02 | 87369 | gtex_brain_ba24 |
| rs13384238 | 2 | 44155986 | LRPPRC | ENSG00000138095.14 | 1.069E-6 | 0.02 | 67158 | gtex_brain_ba24 |
| rs4507144 | 2 | 44164481 | LRPPRC | ENSG00000138095.14 | 1.172E-6 | 0.02 | 58663 | gtex_brain_ba24 |
| rs9309111 | 2 | 44165733 | LRPPRC | ENSG00000138095.14 | 1.301E-6 | 0.02 | 57411 | gtex_brain_ba24 |
| rs28394191 | 2 | 44172444 | LRPPRC | ENSG00000138095.14 | 1.948E-6 | 0.02 | 50700 | gtex_brain_ba24 |
| rs9309111 | 2 | 44165733 | LRPPRC | ENSG00000138095.14 | 1.647E-6 | 0 | 57411 | gtex_brain_putamen_basal |
| rs7594526 | 2 | 44173422 | LRPPRC | ENSG00000138095.14 | 1.244E-6 | 0 | 49722 | gtex_brain_putamen_basal |
| rs200897097 | 2 | 44209046 | LRPPRC | ENSG00000138095.14 | 1.775E-6 | 0 | 14098 | gtex_brain_putamen_basal |
| rs6752841 | 2 | 44209048 | LRPPRC | ENSG00000138095.14 | 1.464E-7 | 0 | 14096 | gtex_brain_putamen_basal |
| rs6753293 | 2 | 44235472 | LRPPRC | ENSG00000138095.14 | 5.928E-7 | 0 | -12328 | gtex_brain_putamen_basal |
| rs6544735 | 2 | 44235780 | LRPPRC | ENSG00000138095.14 | 8.664E-7 | 0 | -12636 | gtex_brain_putamen_basal |
| rs6544736 | 2 | 44236967 | LRPPRC | ENSG00000138095.14 | 7.778E-7 | 0 | -13823 | gtex_brain_putamen_basal |
| rs9678931 | 2 | 44237335 | LRPPRC | ENSG00000138095.14 | 5.261E-7 | 0 | -14191 | gtex_brain_putamen_basal |
| rs4131367 | 2 | 44237874 | LRPPRC | ENSG00000138095.14 | 3.108E-7 | 0 | -14730 | gtex_brain_putamen_basal |
| rs10203839 | 2 | 44239095 | LRPPRC | ENSG00000138095.14 | 2.966E-7 | 0 | -15951 | gtex_brain_putamen_basal |
| rs10186873 | 2 | 44241147 | LRPPRC | ENSG00000138095.14 | 2.522E-6 | 0 | -18003 | gtex_brain_putamen_basal |
| rs11687002 | 2 | 44248383 | LRPPRC | ENSG00000138095.14 | 2.429E-6 | 0 | -25239 | gtex_brain_putamen_basal |
| rs11683522 | 2 | 44249687 | LRPPRC | ENSG00000138095.14 | 3.02E-6 | 0 | -26543 | gtex_brain_putamen_basal |
| rs11683574 | 2 | 44249811 | LRPPRC | ENSG00000138095.14 | 9.302E-7 | 0 | -26667 | gtex_brain_putamen_basal |
| rs11684838 | 2 | 44250155 | LRPPRC | ENSG00000138095.14 | 1.685E-6 | 0 | -27011 | gtex_brain_putamen_basal |
| rs11688286 | 2 | 44252765 | LRPPRC | ENSG00000138095.14 | 2.024E-6 | 0 | -29621 | gtex_brain_putamen_basal |
| rs13007140 | 2 | 44258817 | LRPPRC | ENSG00000138095.14 | 4.28E-7 | 0 | -35673 | gtex_brain_putamen_basal |
| rs7598793 | 2 | 44259490 | LRPPRC | ENSG00000138095.14 | 1.928E-6 | 0 | -36346 | gtex_brain_putamen_basal |
| rs7598912 | 2 | 44259626 | LRPPRC | ENSG00000138095.14 | 1.077E-6 | 0 | -36482 | gtex_brain_putamen_basal |
| rs6744602 | 2 | 44261185 | LRPPRC | ENSG00000138095.14 | 2.652E-6 | 0 | -38041 | gtex_brain_putamen_basal |
| rs3853704 | 2 | 44261528 | LRPPRC | ENSG00000138095.14 | 1.308E-6 | 0 | -38384 | gtex_brain_putamen_basal |
Section II. Transcriptome annotation
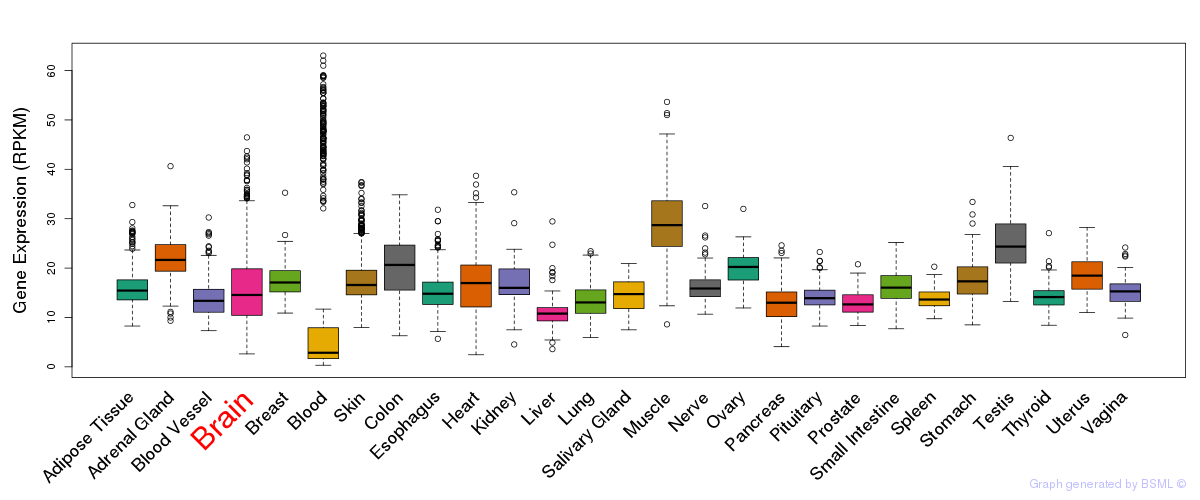
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| PHF6 | 0.97 | 0.94 |
| PRPF40A | 0.97 | 0.96 |
| FAM76B | 0.97 | 0.95 |
| MSH6 | 0.96 | 0.96 |
| PDCL | 0.96 | 0.93 |
| NUP205 | 0.96 | 0.95 |
| PHF16 | 0.96 | 0.93 |
| SMC3 | 0.96 | 0.96 |
| DHX9 | 0.96 | 0.94 |
| SP3 | 0.96 | 0.95 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| HLA-F | -0.68 | -0.77 |
| C5orf53 | -0.67 | -0.73 |
| PTH1R | -0.66 | -0.75 |
| AIFM3 | -0.66 | -0.74 |
| TNFSF12 | -0.66 | -0.68 |
| AF347015.31 | -0.66 | -0.86 |
| AF347015.27 | -0.65 | -0.84 |
| FBXO2 | -0.65 | -0.66 |
| FXYD1 | -0.65 | -0.86 |
| MT-CO2 | -0.65 | -0.87 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003677 | DNA binding | IEA | - | |
| GO:0003723 | RNA binding | NAS | 12832482 | |
| GO:0005515 | protein binding | IPI | 17050673 |17353931 | |
| GO:0008017 | microtubule binding | TAS | 12762840 | |
| GO:0051015 | actin filament binding | IDA | 12762840 | |
| GO:0048487 | beta-tubulin binding | IDA | 12762840 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006355 | regulation of transcription, DNA-dependent | IEA | - | |
| GO:0006350 | transcription | IEA | - | |
| GO:0006810 | transport | IEA | - | |
| GO:0051028 | mRNA transport | IEA | - | |
| GO:0047497 | mitochondrion transport along microtubule | TAS | 12762840 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0000794 | condensed nuclear chromosome | IDA | 12762840 | |
| GO:0005856 | cytoskeleton | IDA | 12762840 | |
| GO:0005634 | nucleus | IDA | 12832482 | |
| GO:0005637 | nuclear inner membrane | IEA | - | |
| GO:0005640 | nuclear outer membrane | IEA | - | |
| GO:0005739 | mitochondrion | IDA | 12832482 | |
| GO:0016020 | membrane | IEA | - | |
| GO:0048471 | perinuclear region of cytoplasm | IDA | 12762840 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ARF6 | DKFZp564M0264 | ADP-ribosylation factor 6 | Affinity Capture-MS | BioGRID | 17353931 |
| CECR2 | KIAA1740 | cat eye syndrome chromosome region, candidate 2 | - | HPRD,BioGRID | 11827465 |
| GABARAPL2 | ATG8 | GATE-16 | GATE16 | GEF-2 | GEF2 | GABA(A) receptor-associated protein-like 2 | Affinity Capture-MS | BioGRID | 17353931 |
| GSTK1 | GST13 | glutathione S-transferase kappa 1 | Affinity Capture-MS | BioGRID | 17353931 |
| HEBP2 | C6ORF34B | C6orf34 | KIAA1244 | PP23 | SOUL | heme binding protein 2 | - | HPRD,BioGRID | 11827465 |
| IKBKE | IKK-i | IKKE | IKKI | KIAA0151 | MGC125294 | MGC125295 | MGC125297 | inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase epsilon | Affinity Capture-MS | BioGRID | 17353931 |
| MAGEA6 | MAGE-3b | MAGE3B | MAGE6 | MGC52297 | melanoma antigen family A, 6 | Affinity Capture-MS | BioGRID | 17353931 |
| MAGED1 | DLXIN-1 | NRAGE | melanoma antigen family D, 1 | Affinity Capture-MS | BioGRID | 17353931 |
| MAP1S | BPY2IP1 | C19orf5 | FLJ10669 | MAP8 | MGC133087 | VCY2IP-1 | VCY2IP1 | microtubule-associated protein 1S | - | HPRD,BioGRID | 11827465 |
| NEK6 | SID6-1512 | NIMA (never in mitosis gene a)-related kinase 6 | Affinity Capture-MS | BioGRID | 17353931 |
| PHLDA3 | TIH1 | pleckstrin homology-like domain, family A, member 3 | Affinity Capture-MS | BioGRID | 17353931 |
| PTP4A3 | PRL-3 | PRL-R | PRL3 | protein tyrosine phosphatase type IVA, member 3 | Affinity Capture-MS | BioGRID | 17353931 |
| TNFRSF10D | CD264 | DCR2 | TRAILR4 | TRUNDD | tumor necrosis factor receptor superfamily, member 10d, decoy with truncated death domain | Affinity Capture-MS | BioGRID | 17353931 |
| UXT | ART-27 | ubiquitously-expressed transcript | - | HPRD,BioGRID | 11827465 |
| WDR8 | FLJ20430 | MGC99569 | WD repeat domain 8 | Affinity Capture-MS | BioGRID | 17353931 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| RHEIN ALL GLUCOCORTICOID THERAPY DN | 362 | 238 | All SZGR 2.0 genes in this pathway |
| TIEN INTESTINE PROBIOTICS 2HR DN | 88 | 53 | All SZGR 2.0 genes in this pathway |
| TIEN INTESTINE PROBIOTICS 6HR DN | 167 | 100 | All SZGR 2.0 genes in this pathway |
| TIEN INTESTINE PROBIOTICS 24HR UP | 557 | 331 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA DN | 1375 | 806 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA POORLY DIFFERENTIATED UP | 633 | 376 | All SZGR 2.0 genes in this pathway |
| OUELLET OVARIAN CANCER INVASIVE VS LMP UP | 117 | 85 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 DN | 855 | 609 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 COMMON DN | 483 | 336 | All SZGR 2.0 genes in this pathway |
| KIM MYC AMPLIFICATION TARGETS UP | 201 | 127 | All SZGR 2.0 genes in this pathway |
| LIAO METASTASIS | 539 | 324 | All SZGR 2.0 genes in this pathway |
| SHIPP DLBCL CURED VS FATAL DN | 45 | 30 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT REJECTED VS OK DN | 546 | 351 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| RODWELL AGING KIDNEY NO BLOOD DN | 150 | 93 | All SZGR 2.0 genes in this pathway |
| RODWELL AGING KIDNEY DN | 145 | 88 | All SZGR 2.0 genes in this pathway |
| BACOLOD RESISTANCE TO ALKYLATING AGENTS DN | 60 | 45 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR UP | 953 | 554 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR DN | 911 | 527 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR DN | 1011 | 592 | All SZGR 2.0 genes in this pathway |
| WONG MITOCHONDRIA GENE MODULE | 217 | 122 | All SZGR 2.0 genes in this pathway |
| BOCHKIS FOXA2 TARGETS | 425 | 261 | All SZGR 2.0 genes in this pathway |
| MOOTHA PGC | 420 | 269 | All SZGR 2.0 genes in this pathway |
| LEE RECENT THYMIC EMIGRANT | 227 | 128 | All SZGR 2.0 genes in this pathway |
| SHEDDEN LUNG CANCER POOR SURVIVAL A6 | 456 | 285 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA CLASSES UP | 605 | 377 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS CALB1 CORR UP | 548 | 370 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS DN | 882 | 538 | All SZGR 2.0 genes in this pathway |
| ROESSLER LIVER CANCER METASTASIS DN | 53 | 29 | All SZGR 2.0 genes in this pathway |