Gene Page: CDH15
Summary ?
| GeneID | 1013 |
| Symbol | CDH15 |
| Synonyms | CDH14|CDH3|CDHM|MCAD|MRD3 |
| Description | cadherin 15 |
| Reference | MIM:114019|HGNC:HGNC:1754|Ensembl:ENSG00000129910|HPRD:00225|Vega:OTTHUMG00000138045 |
| Gene type | protein-coding |
| Map location | 16q24.3 |
| Pascal p-value | 0.002 |
| Fetal beta | -0.015 |
| eGene | Cerebellum Myers' cis & trans |
| Support | CELL ADHESION AND TRANSSYNAPTIC SIGNALING |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs17392743 | chr1 | 54764240 | CDH15 | 1013 | 0.15 | trans | ||
| rs2502827 | chr1 | 176044216 | CDH15 | 1013 | 0.12 | trans | ||
| rs12405921 | chr1 | 206695730 | CDH15 | 1013 | 0.07 | trans | ||
| rs17572651 | chr1 | 218943612 | CDH15 | 1013 | 0 | trans | ||
| rs16829545 | chr2 | 151977407 | CDH15 | 1013 | 3.942E-21 | trans | ||
| rs16841750 | chr2 | 158288461 | CDH15 | 1013 | 0.15 | trans | ||
| rs3845734 | chr2 | 171125572 | CDH15 | 1013 | 1.978E-4 | trans | ||
| rs7584986 | chr2 | 184111432 | CDH15 | 1013 | 2.859E-10 | trans | ||
| rs2183142 | chr4 | 159232695 | CDH15 | 1013 | 0.02 | trans | ||
| rs1396222 | chr4 | 173279496 | CDH15 | 1013 | 0.14 | trans | ||
| rs335980 | chr4 | 173329784 | CDH15 | 1013 | 0.07 | trans | ||
| rs337984 | chr4 | 173411662 | CDH15 | 1013 | 0.15 | trans | ||
| rs17762315 | chr5 | 76807576 | CDH15 | 1013 | 5.54E-4 | trans | ||
| rs7729096 | chr5 | 76835927 | CDH15 | 1013 | 0.16 | trans | ||
| rs9461864 | chr6 | 33481468 | CDH15 | 1013 | 0.06 | trans | ||
| rs3118341 | chr9 | 25185518 | CDH15 | 1013 | 0.05 | trans | ||
| rs11139334 | chr9 | 84209393 | CDH15 | 1013 | 0.01 | trans | ||
| rs2393316 | chr10 | 59333070 | CDH15 | 1013 | 0 | trans | ||
| rs1526959 | chr12 | 79753789 | CDH15 | 1013 | 0.11 | trans | ||
| rs16955618 | chr15 | 29937543 | CDH15 | 1013 | 3.157E-33 | trans | ||
| rs16945437 | chr17 | 11797650 | CDH15 | 1013 | 0.18 | trans | ||
| rs11873184 | chr18 | 1584081 | CDH15 | 1013 | 0.09 | trans | ||
| rs1041786 | chr21 | 22617710 | CDH15 | 1013 | 1.155E-5 | trans | ||
| rs16986332 | chrX | 10958354 | CDH15 | 1013 | 0.06 | trans |
Section II. Transcriptome annotation
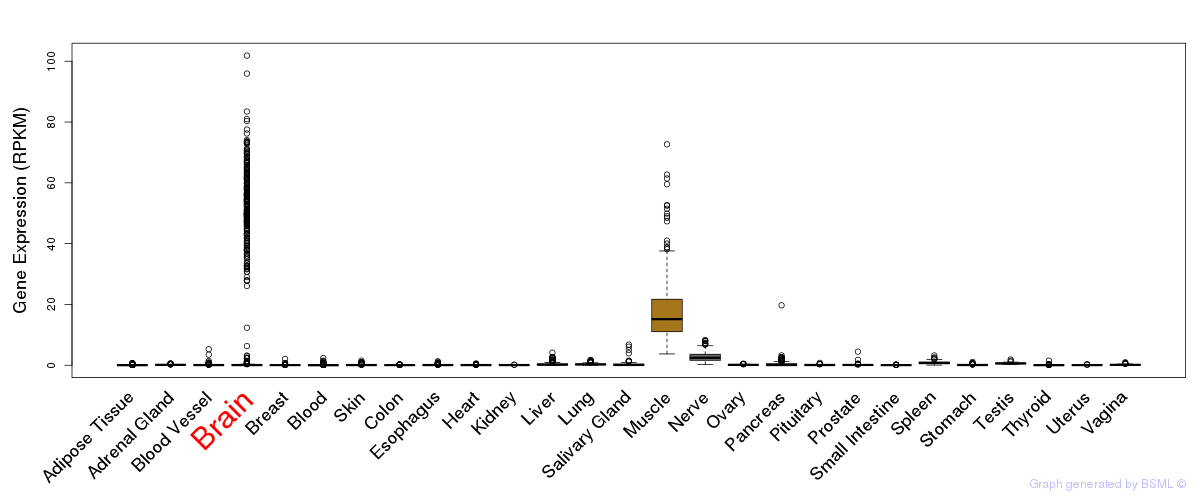
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG CELL ADHESION MOLECULES CAMS | 134 | 93 | All SZGR 2.0 genes in this pathway |
| REACTOME DEVELOPMENTAL BIOLOGY | 396 | 292 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CELL COMMUNICATION | 120 | 77 | All SZGR 2.0 genes in this pathway |
| REACTOME ADHERENS JUNCTIONS INTERACTIONS | 27 | 20 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CELL JUNCTION ORGANIZATION | 56 | 31 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL JUNCTION ORGANIZATION | 78 | 43 | All SZGR 2.0 genes in this pathway |
| REACTOME MYOGENESIS | 28 | 20 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE LIVE UP | 485 | 293 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE LPS UP | 431 | 237 | All SZGR 2.0 genes in this pathway |
| LOPEZ MBD TARGETS | 957 | 597 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY BREAST CANCER 16Q24 AMPLICON | 53 | 38 | All SZGR 2.0 genes in this pathway |
| JECHLINGER EPITHELIAL TO MESENCHYMAL TRANSITION UP | 71 | 51 | All SZGR 2.0 genes in this pathway |
| TARTE PLASMA CELL VS PLASMABLAST UP | 398 | 262 | All SZGR 2.0 genes in this pathway |
| REN ALVEOLAR RHABDOMYOSARCOMA UP | 98 | 64 | All SZGR 2.0 genes in this pathway |
| HU GENOTOXIC DAMAGE 24HR | 35 | 22 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS STEM CELL AND PROGENITOR | 681 | 420 | All SZGR 2.0 genes in this pathway |
| YOSHIOKA LIVER CANCER EARLY RECURRENCE DN | 65 | 31 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN ES ICP WITH H3K4ME3 | 718 | 401 | All SZGR 2.0 genes in this pathway |