Gene Page: CDK7
Summary ?
| GeneID | 1022 |
| Symbol | CDK7 |
| Synonyms | CAK1|CDKN7|HCAK|MO15|STK1|p39MO15 |
| Description | cyclin-dependent kinase 7 |
| Reference | MIM:601955|HGNC:HGNC:1778|Ensembl:ENSG00000134058|HPRD:15993|Vega:OTTHUMG00000099358 |
| Gene type | protein-coding |
| Map location | 5q12.1 |
| Pascal p-value | 0.844 |
| Sherlock p-value | 0.011 |
| Fetal beta | 0.48 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
General gene expression (GTEx)
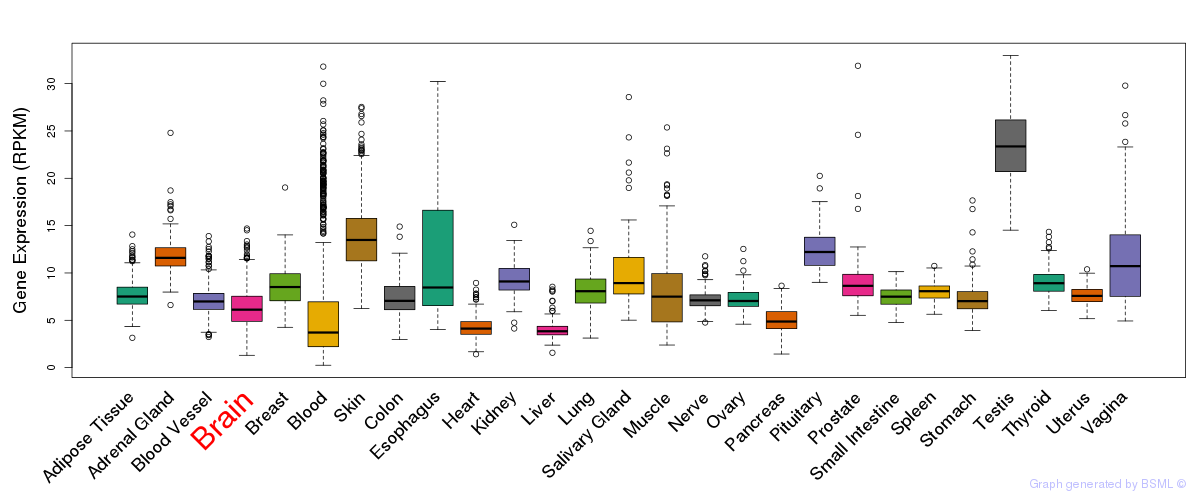
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| BOK | 0.80 | 0.82 |
| SLC26A11 | 0.75 | 0.70 |
| VAT1 | 0.74 | 0.67 |
| RGS19 | 0.73 | 0.58 |
| EPS8L1 | 0.71 | 0.41 |
| RAC3 | 0.71 | 0.52 |
| MVK | 0.71 | 0.73 |
| PIP5KL1 | 0.69 | 0.51 |
| NAGLU | 0.69 | 0.70 |
| RNF220 | 0.68 | 0.57 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.31 | -0.45 | -0.57 |
| MT-CO2 | -0.44 | -0.57 |
| AF347015.8 | -0.42 | -0.56 |
| AF347015.21 | -0.42 | -0.60 |
| MT-CYB | -0.41 | -0.55 |
| CXCL14 | -0.41 | -0.53 |
| AF347015.27 | -0.41 | -0.53 |
| CARD16 | -0.40 | -0.58 |
| AF347015.33 | -0.40 | -0.51 |
| EAF2 | -0.40 | -0.51 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0004693 | cyclin-dependent protein kinase activity | TAS | neuron (GO term level: 8) | 8208544 |
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0003713 | transcription coactivator activity | NAS | 15572661 | |
| GO:0005524 | ATP binding | IEA | - | |
| GO:0016740 | transferase activity | IEA | - | |
| GO:0008353 | RNA polymerase subunit kinase activity | IEA | - | |
| GO:0008022 | protein C-terminus binding | IPI | 10801852 | |
| GO:0050681 | androgen receptor binding | NAS | 15572661 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0000079 | regulation of cyclin-dependent protein kinase activity | TAS | 7533895 | |
| GO:0000718 | nucleotide-excision repair, DNA damage removal | EXP | 10583946 | |
| GO:0006367 | transcription initiation from RNA polymerase II promoter | EXP | 8946909 | |
| GO:0006367 | transcription initiation from RNA polymerase II promoter | TAS | 7533895 | |
| GO:0006368 | RNA elongation from RNA polymerase II promoter | EXP | 9405375 | |
| GO:0006350 | transcription | IEA | - | |
| GO:0006468 | protein amino acid phosphorylation | IEA | - | |
| GO:0006281 | DNA repair | IEA | - | |
| GO:0007049 | cell cycle | IEA | - | |
| GO:0008283 | cell proliferation | TAS | 7533895 | |
| GO:0051301 | cell division | IEA | - | |
| GO:0030521 | androgen receptor signaling pathway | NAS | 15572661 | |
| GO:0045944 | positive regulation of transcription from RNA polymerase II promoter | IDA | 8692841 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005634 | nucleus | IDA | 16109376 | |
| GO:0005654 | nucleoplasm | EXP | 1939271 |2449431 |7629134 |7799941 |8946909 |9512541 |9582279 |9790902 |10214908 |11313499 |12393749 |12646563 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| AR | AIS | DHTR | HUMARA | KD | NR3C4 | SBMA | SMAX1 | TFM | androgen receptor | Affinity Capture-Western Reconstituted Complex | BioGRID | 10734072 |
| CCNH | CAK | p34 | p37 | cyclin H | Affinity Capture-MS Affinity Capture-Western Two-hybrid | BioGRID | 8078587 |
| CCNH | CAK | p34 | p37 | cyclin H | - | HPRD | 9056480 |
| CDK2 | p33(CDK2) | cyclin-dependent kinase 2 | CDK7 phosphorylates an unspecified isoform of CDK2. | BIND | 7629134 |
| CDK6 | MGC59692 | PLSTIRE | STQTL11 | cyclin-dependent kinase 6 | CAK phosphorylates CDK6. This interaction was modeled on a demonstrated interaction between human CAK and CDK6 from an unspecified species. | BIND | 7629134 |
| CUX1 | CASP | CDP | CDP/Cut | CDP1 | COY1 | CUTL1 | CUX | Clox | Cux/CDP | GOLIM6 | Nbla10317 | p100 | p110 | p200 | p75 | cut-like homeobox 1 | Reconstituted Complex | BioGRID | 11584018 |
| E2F1 | E2F-1 | RBAP1 | RBBP3 | RBP3 | E2F transcription factor 1 | Affinity Capture-Western | BioGRID | 10428966 |
| ERCC2 | COFS2 | EM9 | MGC102762 | MGC126218 | MGC126219 | TTD | XPD | excision repair cross-complementing rodent repair deficiency, complementation group 2 | - | HPRD | 11445587 |
| ERCC3 | BTF2 | GTF2H | RAD25 | TFIIH | XPB | excision repair cross-complementing rodent repair deficiency, complementation group 3 (xeroderma pigmentosum group B complementing) | - | HPRD,BioGRID | 9130708 |
| ESR1 | DKFZp686N23123 | ER | ESR | ESRA | Era | NR3A1 | estrogen receptor 1 | Biochemical Activity | BioGRID | 10949034 |
| GTF2H1 | BTF2 | TFB1 | TFIIH | general transcription factor IIH, polypeptide 1, 62kDa | - | HPRD,BioGRID | 9130708 |
| GTF2H2 | BTF2 | BTF2P44 | MGC102806 | T-BTF2P44 | TFIIH | general transcription factor IIH, polypeptide 2, 44kDa | - | HPRD | 9130708 |
| GTF2H5 | C6orf175 | TFB5 | TGF2H5 | TTD | TTD-A | TTDA | bA120J8.2 | general transcription factor IIH, polypeptide 5 | - | HPRD,BioGRID | 11062469 |15220921 |
| HDAC2 | RPD3 | YAF1 | histone deacetylase 2 | Affinity Capture-Western | BioGRID | 12527756 |
| HNRNPU | HNRPU | SAF-A | U21.1 | heterogeneous nuclear ribonucleoprotein U (scaffold attachment factor A) | Affinity Capture-Western | BioGRID | 10490622 |
| MCM7 | CDABP0042 | CDC47 | MCM2 | P1.1-MCM3 | P1CDC47 | P85MCM | PNAS-146 | minichromosome maintenance complex component 7 | - | HPRD,BioGRID | 11056214 |
| MED21 | SRB7 | SURB7 | mediator complex subunit 21 | Affinity Capture-Western | BioGRID | 9315662 |
| MNAT1 | MAT1 | RNF66 | TFB3 | menage a trois homolog 1, cyclin H assembly factor (Xenopus laevis) | - | HPRD,BioGRID | 8521393 |
| MNAT1 | MAT1 | RNF66 | TFB3 | menage a trois homolog 1, cyclin H assembly factor (Xenopus laevis) | - | HPRD | 9130708 |
| MTA1 | - | metastasis associated 1 | Affinity Capture-Western | BioGRID | 12527756 |
| RARA | NR1B1 | RAR | retinoic acid receptor, alpha | RAR-alpha interacts with hMo15. | BIND | 15249124 |
| TCEA1 | GTF2S | SII | TCEA | TF2S | TFIIS | transcription elongation factor A (SII), 1 | Reconstituted Complex | BioGRID | 9305922 |
| THRA | AR7 | EAR7 | ERB-T-1 | ERBA | ERBA1 | MGC000261 | MGC43240 | NR1A1 | THRA1 | THRA2 | c-ERBA-1 | thyroid hormone receptor, alpha (erythroblastic leukemia viral (v-erb-a) oncogene homolog, avian) | TR-alpha interacts with hMo15. This interaction was modeled on a demonstrated interaction between chicken TR-alpha and human hMo15. | BIND | 15249124 |
| TP53 | FLJ92943 | LFS1 | TRP53 | p53 | tumor protein p53 | Affinity Capture-Western Biochemical Activity | BioGRID | 9372954 |9840937 |
| VDR | NR1I1 | vitamin D (1,25- dihydroxyvitamin D3) receptor | VDR interacts with hMo15. | BIND | 15249124 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG NUCLEOTIDE EXCISION REPAIR | 44 | 25 | All SZGR 2.0 genes in this pathway |
| KEGG CELL CYCLE | 128 | 84 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CELLCYCLE PATHWAY | 23 | 15 | All SZGR 2.0 genes in this pathway |
| BIOCARTA PTC1 PATHWAY | 11 | 6 | All SZGR 2.0 genes in this pathway |
| SA G2 AND M PHASES | 8 | 7 | All SZGR 2.0 genes in this pathway |
| PID RETINOIC ACID PATHWAY | 30 | 23 | All SZGR 2.0 genes in this pathway |
| REACTOME RNA POL I TRANSCRIPTION TERMINATION | 22 | 12 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CYCLE | 421 | 253 | All SZGR 2.0 genes in this pathway |
| REACTOME RNA POL I TRANSCRIPTION | 89 | 64 | All SZGR 2.0 genes in this pathway |
| REACTOME RNA POL II TRANSCRIPTION | 105 | 54 | All SZGR 2.0 genes in this pathway |
| REACTOME MRNA CAPPING | 30 | 11 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CYCLE MITOTIC | 325 | 185 | All SZGR 2.0 genes in this pathway |
| REACTOME CYCLIN E ASSOCIATED EVENTS DURING G1 S TRANSITION | 65 | 40 | All SZGR 2.0 genes in this pathway |
| REACTOME G1 PHASE | 38 | 23 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSCRIPTION COUPLED NER TC NER | 45 | 24 | All SZGR 2.0 genes in this pathway |
| REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | 41 | 21 | All SZGR 2.0 genes in this pathway |
| REACTOME MRNA PROCESSING | 161 | 86 | All SZGR 2.0 genes in this pathway |
| REACTOME G1 S TRANSITION | 112 | 63 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSCRIPTION | 210 | 127 | All SZGR 2.0 genes in this pathway |
| REACTOME NUCLEOTIDE EXCISION REPAIR | 51 | 27 | All SZGR 2.0 genes in this pathway |
| REACTOME FORMATION OF RNA POL II ELONGATION COMPLEX | 45 | 19 | All SZGR 2.0 genes in this pathway |
| REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | 15 | 8 | All SZGR 2.0 genes in this pathway |
| REACTOME FORMATION OF TRANSCRIPTION COUPLED NER TC NER REPAIR COMPLEX | 30 | 13 | All SZGR 2.0 genes in this pathway |
| REACTOME MITOTIC G1 G1 S PHASES | 137 | 79 | All SZGR 2.0 genes in this pathway |
| REACTOME RNA POL I RNA POL III AND MITOCHONDRIAL TRANSCRIPTION | 122 | 80 | All SZGR 2.0 genes in this pathway |
| REACTOME MITOTIC G2 G2 M PHASES | 81 | 50 | All SZGR 2.0 genes in this pathway |
| REACTOME DNA REPAIR | 112 | 59 | All SZGR 2.0 genes in this pathway |
| REACTOME RNA POL II PRE TRANSCRIPTION EVENTS | 61 | 32 | All SZGR 2.0 genes in this pathway |
| REACTOME GLOBAL GENOMIC NER GG NER | 35 | 20 | All SZGR 2.0 genes in this pathway |
| REACTOME FORMATION OF INCISION COMPLEX IN GG NER | 23 | 11 | All SZGR 2.0 genes in this pathway |
| REACTOME HIV INFECTION | 207 | 122 | All SZGR 2.0 genes in this pathway |
| REACTOME HIV LIFE CYCLE | 125 | 69 | All SZGR 2.0 genes in this pathway |
| REACTOME FORMATION OF THE HIV1 EARLY ELONGATION COMPLEX | 34 | 11 | All SZGR 2.0 genes in this pathway |
| REACTOME LATE PHASE OF HIV LIFE CYCLE | 104 | 61 | All SZGR 2.0 genes in this pathway |
| REACTOME S PHASE | 109 | 66 | All SZGR 2.0 genes in this pathway |
| REACTOME RNA POL I TRANSCRIPTION INITIATION | 25 | 14 | All SZGR 2.0 genes in this pathway |
| PUIFFE INVASION INHIBITED BY ASCITES DN | 145 | 91 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| BORCZUK MALIGNANT MESOTHELIOMA UP | 305 | 185 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA ANAPLASTIC UP | 722 | 443 | All SZGR 2.0 genes in this pathway |
| HAHTOLA SEZARY SYNDROM UP | 98 | 58 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE EPIDERMIS DN | 508 | 354 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 3 DN | 229 | 142 | All SZGR 2.0 genes in this pathway |
| WANG ESOPHAGUS CANCER VS NORMAL DN | 101 | 66 | All SZGR 2.0 genes in this pathway |
| SILIGAN TARGETS OF EWS FLI1 FUSION UP | 15 | 12 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER SERUM RESPONSE DN | 712 | 443 | All SZGR 2.0 genes in this pathway |
| PATIL LIVER CANCER | 747 | 453 | All SZGR 2.0 genes in this pathway |
| GRUETZMANN PANCREATIC CANCER UP | 358 | 245 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS DN | 848 | 527 | All SZGR 2.0 genes in this pathway |
| BENPORATH CYCLING GENES | 648 | 385 | All SZGR 2.0 genes in this pathway |
| SAKAI TUMOR INFILTRATING MONOCYTES DN | 81 | 51 | All SZGR 2.0 genes in this pathway |
| KAUFFMANN DNA REPAIR GENES | 230 | 137 | All SZGR 2.0 genes in this pathway |
| DORSAM HOXA9 TARGETS UP | 35 | 25 | All SZGR 2.0 genes in this pathway |
| FLECHNER PBL KIDNEY TRANSPLANT REJECTED VS OK UP | 63 | 48 | All SZGR 2.0 genes in this pathway |
| KANG IMMORTALIZED BY TERT UP | 89 | 61 | All SZGR 2.0 genes in this pathway |
| ZHAN V2 LATE DIFFERENTIATION GENES | 45 | 34 | All SZGR 2.0 genes in this pathway |
| ZAMORA NOS2 TARGETS UP | 69 | 41 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| TAKAO RESPONSE TO UVB RADIATION DN | 98 | 67 | All SZGR 2.0 genes in this pathway |
| GENTILE UV RESPONSE CLUSTER D6 | 37 | 25 | All SZGR 2.0 genes in this pathway |
| GENTILE UV HIGH DOSE DN | 312 | 203 | All SZGR 2.0 genes in this pathway |
| SESTO RESPONSE TO UV C0 | 107 | 72 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE INCIPIENT DN | 165 | 106 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RESPONSE TO TRABECTEDIN DN | 271 | 175 | All SZGR 2.0 genes in this pathway |
| MONNIER POSTRADIATION TUMOR ESCAPE DN | 373 | 196 | All SZGR 2.0 genes in this pathway |
| GRADE COLON CANCER UP | 871 | 505 | All SZGR 2.0 genes in this pathway |
| GRADE COLON AND RECTAL CANCER UP | 285 | 167 | All SZGR 2.0 genes in this pathway |
| BOQUEST STEM CELL CULTURED VS FRESH UP | 425 | 298 | All SZGR 2.0 genes in this pathway |
| CHEN HOXA5 TARGETS 9HR UP | 223 | 132 | All SZGR 2.0 genes in this pathway |
| BLUM RESPONSE TO SALIRASIB UP | 245 | 159 | All SZGR 2.0 genes in this pathway |
| MUELLER PLURINET | 299 | 189 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS DN | 366 | 257 | All SZGR 2.0 genes in this pathway |
| WHITFIELD CELL CYCLE M G1 | 148 | 95 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS DN | 882 | 538 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION DN | 1080 | 713 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-496 | 114 | 121 | 1A,m8 | hsa-miR-496 | AUUACAUGGCCAAUCUC |
| hsa-miR-496 | AUUACAUGGCCAAUCUC | ||||
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.