Gene Page: SRRM1
Summary ?
| GeneID | 10250 |
| Symbol | SRRM1 |
| Synonyms | 160-KD|POP101|SRM160 |
| Description | serine and arginine repetitive matrix 1 |
| Reference | MIM:605975|HGNC:HGNC:16638|Ensembl:ENSG00000133226|HPRD:10441|Vega:OTTHUMG00000003320 |
| Gene type | protein-coding |
| Map location | 1p36.11 |
| Pascal p-value | 0.024 |
| Sherlock p-value | 0.892 |
| Fetal beta | 0.575 |
| DMG | 1 (# studies) |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg12657664 | 1 | 24969003 | SRRM1 | 6.08E-8 | -0.015 | 1.53E-5 | DMG:Jaffe_2016 |
Section II. Transcriptome annotation
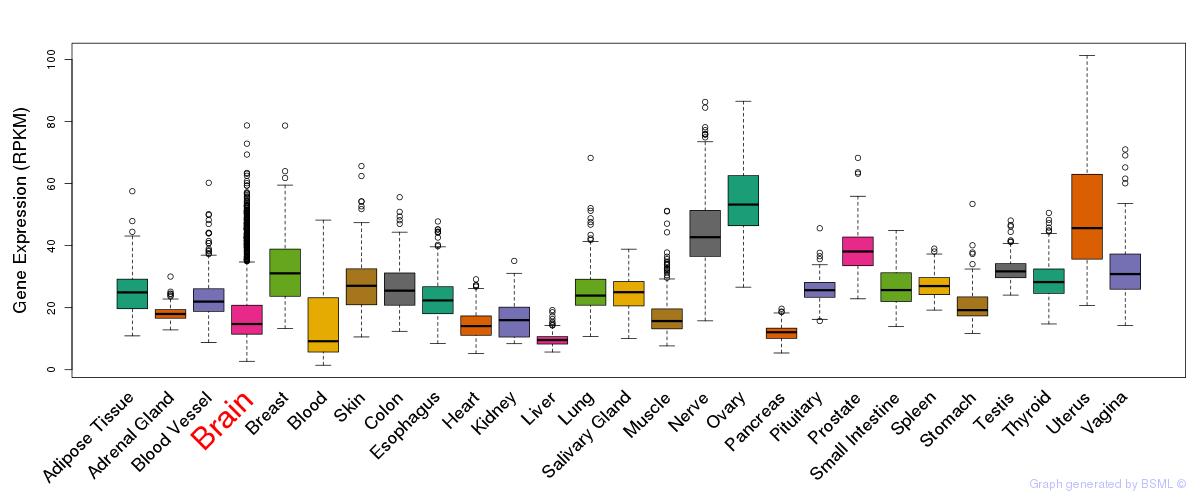
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| FAM124A | 0.85 | 0.84 |
| GAB2 | 0.82 | 0.83 |
| SCD5 | 0.81 | 0.77 |
| ZDHHC9 | 0.76 | 0.77 |
| UNC84B | 0.76 | 0.79 |
| FAM123A | 0.76 | 0.74 |
| RP1-21O18.1 | 0.76 | 0.77 |
| SSBP3 | 0.75 | 0.72 |
| TMEM108 | 0.74 | 0.58 |
| LIMCH1 | 0.74 | 0.84 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AC007405.8 | -0.57 | -0.61 |
| DBI | -0.56 | -0.63 |
| EAF2 | -0.55 | -0.58 |
| S100A16 | -0.54 | -0.58 |
| GNG11 | -0.53 | -0.59 |
| AF347015.21 | -0.52 | -0.60 |
| CXCL14 | -0.52 | -0.58 |
| FGGY | -0.52 | -0.56 |
| HSD17B14 | -0.51 | -0.57 |
| AF347015.31 | -0.51 | -0.54 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| DEK | D6S231E | DEK oncogene | - | HPRD,BioGRID | 10908574 |
| FUS | CHOP | FUS-CHOP | FUS1 | TLS | TLS/CHOP | hnRNP-P2 | fusion (involved in t(12;16) in malignant liposarcoma) | - | HPRD,BioGRID | 12581738 |
| NXF1 | DKFZp667O0311 | MEX67 | TAP | nuclear RNA export factor 1 | - | HPRD,BioGRID | 12093754 |
| RBM8A | BOV-1A | BOV-1B | BOV-1C | MDS014 | RBM8 | RBM8B | Y14 | ZNRP | ZRNP1 | RNA binding motif protein 8A | - | HPRD,BioGRID | 12093754 |
| RNPS1 | E5.1 | MGC117332 | RNA binding protein S1, serine-rich domain | - | HPRD,BioGRID | 12093754 |
| SFRS4 | SRP75 | splicing factor, arginine/serine-rich 4 | - | HPRD,BioGRID | 9531537 |
| SFRS5 | HRS | SRP40 | splicing factor, arginine/serine-rich 5 | - | HPRD,BioGRID | 9531537 |
| SRRM2 | 300-KD | CWF21 | DKFZp686O15166 | FLJ21926 | FLJ22250 | KIAA0324 | MGC40295 | SRL300 | SRm300 | serine/arginine repetitive matrix 2 | - | HPRD,BioGRID | 9531537 |
| UPF2 | DKFZp434D222 | HUPF2 | KIAA1408 | MGC138834 | MGC138835 | RENT2 | smg-3 | UPF2 regulator of nonsense transcripts homolog (yeast) | - | HPRD,BioGRID | 12093754 |
| UPF3B | HUPF3B | MRXS14 | RENT3B | UPF3X | UPF3 regulator of nonsense transcripts homolog B (yeast) | - | HPRD,BioGRID | 12093754 |
| YWHAG | 14-3-3GAMMA | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma polypeptide | - | HPRD | 15324660 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| REACTOME PROCESSING OF CAPPED INTRON CONTAINING PRE MRNA | 140 | 77 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSPORT OF MATURE TRANSCRIPT TO CYTOPLASM | 54 | 34 | All SZGR 2.0 genes in this pathway |
| REACTOME RNA POL II TRANSCRIPTION | 105 | 54 | All SZGR 2.0 genes in this pathway |
| REACTOME MRNA PROCESSING | 161 | 86 | All SZGR 2.0 genes in this pathway |
| REACTOME MRNA SPLICING | 111 | 58 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSCRIPTION | 210 | 127 | All SZGR 2.0 genes in this pathway |
| REACTOME MRNA 3 END PROCESSING | 35 | 18 | All SZGR 2.0 genes in this pathway |
| REACTOME CLEAVAGE OF GROWING TRANSCRIPT IN THE TERMINATION REGION | 44 | 22 | All SZGR 2.0 genes in this pathway |
| LIU SOX4 TARGETS DN | 309 | 191 | All SZGR 2.0 genes in this pathway |
| PUIFFE INVASION INHIBITED BY ASCITES UP | 82 | 51 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| WANG CLIM2 TARGETS DN | 186 | 114 | All SZGR 2.0 genes in this pathway |
| OSWALD HEMATOPOIETIC STEM CELL IN COLLAGEN GEL UP | 233 | 161 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION UP | 1278 | 748 | All SZGR 2.0 genes in this pathway |
| MAYBURD RESPONSE TO L663536 DN | 56 | 32 | All SZGR 2.0 genes in this pathway |
| HAHTOLA MYCOSIS FUNGOIDES SKIN UP | 177 | 113 | All SZGR 2.0 genes in this pathway |
| LASTOWSKA NEUROBLASTOMA COPY NUMBER DN | 800 | 473 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER SERUM RESPONSE DN | 712 | 443 | All SZGR 2.0 genes in this pathway |
| WOOD EBV EBNA1 TARGETS UP | 110 | 71 | All SZGR 2.0 genes in this pathway |
| JAZAG TGFB1 SIGNALING VIA SMAD4 UP | 108 | 66 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| GARCIA TARGETS OF FLI1 AND DAX1 DN | 176 | 104 | All SZGR 2.0 genes in this pathway |
| STARK PREFRONTAL CORTEX 22Q11 DELETION UP | 195 | 138 | All SZGR 2.0 genes in this pathway |
| FLECHNER PBL KIDNEY TRANSPLANT OK VS DONOR UP | 151 | 100 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL QTL TRANS | 882 | 572 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT REJECTED VS OK DN | 546 | 351 | All SZGR 2.0 genes in this pathway |
| PENG GLUTAMINE DEPRIVATION DN | 337 | 230 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT OK VS DONOR UP | 555 | 346 | All SZGR 2.0 genes in this pathway |
| STONER ESOPHAGEAL CARCINOGENESIS DN | 7 | 6 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 TARGETS UP | 673 | 430 | All SZGR 2.0 genes in this pathway |
| MARTINEZ TP53 TARGETS DN | 593 | 372 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 AND TP53 TARGETS DN | 591 | 366 | All SZGR 2.0 genes in this pathway |
| RIZKI TUMOR INVASIVENESS 2D UP | 69 | 46 | All SZGR 2.0 genes in this pathway |
| WANG TUMOR INVASIVENESS DN | 210 | 128 | All SZGR 2.0 genes in this pathway |
| AGUIRRE PANCREATIC CANCER COPY NUMBER DN | 238 | 145 | All SZGR 2.0 genes in this pathway |
| SHEDDEN LUNG CANCER GOOD SURVIVAL A12 | 317 | 177 | All SZGR 2.0 genes in this pathway |
| FOURNIER ACINAR DEVELOPMENT LATE 2 | 277 | 172 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 2 DN | 336 | 211 | All SZGR 2.0 genes in this pathway |
| FEVR CTNNB1 TARGETS DN | 553 | 343 | All SZGR 2.0 genes in this pathway |