Gene Page: SMNDC1
Summary ?
| GeneID | 10285 |
| Symbol | SMNDC1 |
| Synonyms | SMNR|SPF30|TDRD16C |
| Description | survival motor neuron domain containing 1 |
| Reference | MIM:603519|HGNC:HGNC:16900|HPRD:04627| |
| Gene type | protein-coding |
| Map location | 10q23 |
| Pascal p-value | 0.447 |
| Sherlock p-value | 0.863 |
| Fetal beta | 0.506 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg21872391 | 10 | 112064057 | SMNDC1 | 1.23E-8 | -0.013 | 5E-6 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs7629431 | chr3 | 189336841 | SMNDC1 | 10285 | 0.14 | trans | ||
| rs4491879 | chr3 | 189373908 | SMNDC1 | 10285 | 0.03 | trans | ||
| rs4505678 | chr3 | 189374356 | SMNDC1 | 10285 | 0.06 | trans |
Section II. Transcriptome annotation
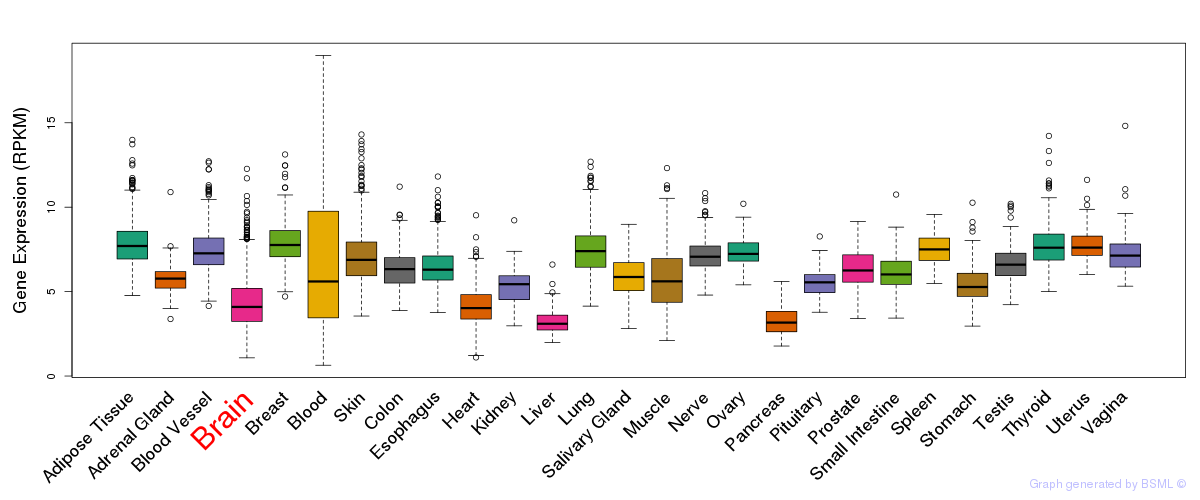
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| PCIF1 | 0.96 | 0.94 |
| ZNF574 | 0.95 | 0.96 |
| ZC3H3 | 0.95 | 0.95 |
| NUP62 | 0.95 | 0.95 |
| GEMIN4 | 0.95 | 0.94 |
| ZNF319 | 0.94 | 0.94 |
| MAPK7 | 0.94 | 0.92 |
| NUP188 | 0.94 | 0.94 |
| ZNF835 | 0.94 | 0.96 |
| USP21 | 0.94 | 0.94 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| C5orf53 | -0.71 | -0.79 |
| AF347015.31 | -0.70 | -0.87 |
| MT-CO2 | -0.69 | -0.88 |
| AF347015.27 | -0.69 | -0.86 |
| AF347015.33 | -0.67 | -0.84 |
| S100B | -0.67 | -0.81 |
| AF347015.8 | -0.66 | -0.86 |
| MT-CYB | -0.66 | -0.83 |
| FXYD1 | -0.65 | -0.81 |
| IFI27 | -0.65 | -0.78 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| CDK5RAP3 | C53 | HSF-27 | IC53 | LZAP | MST016 | OK/SW-cl.114 | CDK5 regulatory subunit associated protein 3 | Affinity Capture-MS | BioGRID | 17353931 |
| DDX21 | DKFZp686F21172 | GUA | GURDB | RH-II/GU | RH-II/GuA | DEAD (Asp-Glu-Ala-Asp) box polypeptide 21 | Affinity Capture-MS | BioGRID | 17353931 |
| EWSR1 | EWS | Ewing sarcoma breakpoint region 1 | Affinity Capture-MS | BioGRID | 17353931 |
| FDFT1 | DGPT | ERG9 | SQS | SS | farnesyl-diphosphate farnesyltransferase 1 | Affinity Capture-MS | BioGRID | 17353931 |
| PPAN | BXDC3 | MGC14226 | MGC45852 | SSF | SSF1 | SSF2 | peter pan homolog (Drosophila) | Affinity Capture-MS | BioGRID | 17353931 |
| RNMTL1 | FLJ10581 | HC90 | RNA methyltransferase like 1 | Affinity Capture-MS | BioGRID | 17353931 |
| RPL3 | MGC104284 | TARBP-B | ribosomal protein L3 | Affinity Capture-MS | BioGRID | 17353931 |
| RUVBL1 | ECP54 | INO80H | NMP238 | PONTIN | Pontin52 | RVB1 | TIH1 | TIP49 | TIP49A | RuvB-like 1 (E. coli) | Affinity Capture-MS | BioGRID | 17353931 |
| SF3A2 | PRP11 | PRPF11 | SAP62 | SF3a66 | splicing factor 3a, subunit 2, 66kDa | Affinity Capture-MS | BioGRID | 12234937 |
| SF3A3 | PRP9 | PRPF9 | SAP61 | SF3a60 | splicing factor 3a, subunit 3, 60kDa | Affinity Capture-MS | BioGRID | 17353931 |
| SF3B1 | PRP10 | PRPF10 | SAP155 | SF3b155 | splicing factor 3b, subunit 1, 155kDa | Affinity Capture-MS | BioGRID | 17353931 |
| SF3B2 | SAP145 | SF3B145 | SF3b1 | SF3b150 | splicing factor 3b, subunit 2, 145kDa | Affinity Capture-MS | BioGRID | 17353931 |
| SF3B3 | KIAA0017 | RSE1 | SAP130 | SF3b130 | STAF130 | splicing factor 3b, subunit 3, 130kDa | Affinity Capture-MS | BioGRID | 17353931 |
| SF3B4 | MGC10828 | SAP49 | SF3b49 | splicing factor 3b, subunit 4, 49kDa | Affinity Capture-MS | BioGRID | 17353931 |
| SNRNP200 | ASCC3L1 | BRR2 | HELIC2 | U5-200KD | small nuclear ribonucleoprotein 200kDa (U5) | - | HPRD,BioGRID | 11331595 |
| TSR1 | FLJ10534 | KIAA1401 | MGC131829 | TSR1, 20S rRNA accumulation, homolog (S. cerevisiae) | Affinity Capture-MS | BioGRID | 17353931 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG SPLICEOSOME | 128 | 72 | All SZGR 2.0 genes in this pathway |
| PRAMOONJAGO SOX4 TARGETS UP | 52 | 38 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| BORCZUK MALIGNANT MESOTHELIOMA UP | 305 | 185 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE EPIDERMIS DN | 508 | 354 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER SERUM RESPONSE DN | 712 | 443 | All SZGR 2.0 genes in this pathway |
| INAMURA LUNG CANCER SCC SUBTYPES UP | 14 | 9 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| PUJANA CHEK2 PCC NETWORK | 779 | 480 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS UP | 769 | 437 | All SZGR 2.0 genes in this pathway |
| WEI MYCN TARGETS WITH E BOX | 795 | 478 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL QTL TRANS | 882 | 572 | All SZGR 2.0 genes in this pathway |
| DEBIASI APOPTOSIS BY REOVIRUS INFECTION UP | 314 | 201 | All SZGR 2.0 genes in this pathway |
| DE YY1 TARGETS DN | 92 | 64 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER UP | 973 | 570 | All SZGR 2.0 genes in this pathway |
| ZHANG BREAST CANCER PROGENITORS UP | 425 | 253 | All SZGR 2.0 genes in this pathway |
| MUELLER PLURINET | 299 | 189 | All SZGR 2.0 genes in this pathway |
| SHEDDEN LUNG CANCER POOR SURVIVAL A6 | 456 | 285 | All SZGR 2.0 genes in this pathway |
| SAKAI CHRONIC HEPATITIS VS LIVER CANCER UP | 83 | 63 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 3 DN | 918 | 550 | All SZGR 2.0 genes in this pathway |