Gene Page: KATNB1
Summary ?
| GeneID | 10300 |
| Symbol | KATNB1 |
| Synonyms | KAT|LIS6 |
| Description | katanin p80 (WD repeat containing) subunit B 1 |
| Reference | MIM:602703|HGNC:HGNC:6217|Ensembl:ENSG00000140854|HPRD:04081|Vega:OTTHUMG00000133467 |
| Gene type | protein-coding |
| Map location | 16q21 |
| Pascal p-value | 0.327 |
| Sherlock p-value | 0.286 |
| Fetal beta | -0.32 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
General gene expression (GTEx)
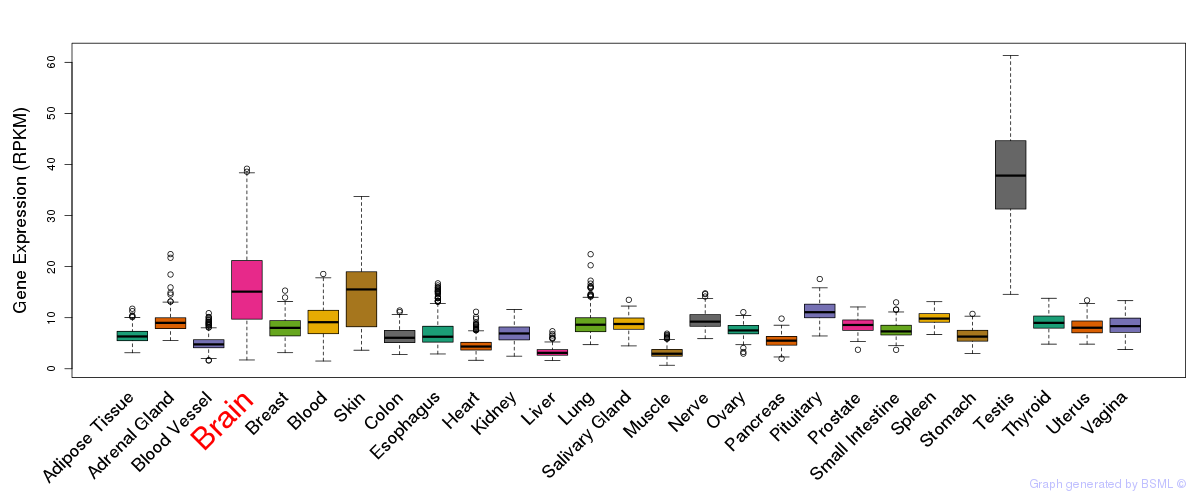
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| DNM1L | 0.84 | 0.88 |
| TRAPPC6B | 0.84 | 0.85 |
| YWHAB | 0.82 | 0.84 |
| USP14 | 0.82 | 0.84 |
| RAB6A | 0.82 | 0.85 |
| NKRF | 0.82 | 0.86 |
| VPS41 | 0.82 | 0.84 |
| SRPK2 | 0.82 | 0.86 |
| NDFIP1 | 0.82 | 0.83 |
| UBE2K | 0.81 | 0.86 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.2 | -0.61 | -0.62 |
| MT-CO2 | -0.60 | -0.63 |
| EIF4EBP3 | -0.60 | -0.67 |
| FXYD1 | -0.60 | -0.62 |
| AF347015.31 | -0.60 | -0.62 |
| ACSF2 | -0.59 | -0.66 |
| AF347015.8 | -0.59 | -0.61 |
| AF347015.21 | -0.58 | -0.57 |
| HIGD1B | -0.57 | -0.62 |
| METRN | -0.57 | -0.65 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0008568 | microtubule-severing ATPase activity | IDA | 10751153 | |
| GO:0008017 | microtubule binding | NAS | 10751153 | |
| GO:0046982 | protein heterodimerization activity | IPI | 10751153 | |
| GO:0045502 | dynein binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007026 | negative regulation of microtubule depolymerization | IMP | 10751153 | |
| GO:0007049 | cell cycle | IEA | - | |
| GO:0007067 | mitosis | IEA | - | |
| GO:0006605 | protein targeting | NAS | 10751153 | |
| GO:0051301 | cell division | IEA | - | |
| GO:0031117 | positive regulation of microtubule depolymerization | IMP | 10751153 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0030424 | axon | IEA | neuron, axon, Neurotransmitter (GO term level: 6) | - |
| GO:0000922 | spindle pole | IDA | 10751153 | |
| GO:0005856 | cytoskeleton | IEA | - | |
| GO:0005874 | microtubule | IEA | - | |
| GO:0005737 | cytoplasm | IDA | 10751153 | |
| GO:0008352 | katanin complex | TAS | 10751153 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| SENGUPTA NASOPHARYNGEAL CARCINOMA DN | 349 | 157 | All SZGR 2.0 genes in this pathway |
| LAIHO COLORECTAL CANCER SERRATED DN | 86 | 47 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 1 DN | 242 | 165 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 2 DN | 281 | 186 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN NPM1 SIGNATURE 3 DN | 162 | 116 | All SZGR 2.0 genes in this pathway |
| UDAYAKUMAR MED1 TARGETS UP | 135 | 82 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC3 TARGETS DN | 536 | 332 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA UP | 1821 | 933 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE KERATINOCYTE DN | 485 | 334 | All SZGR 2.0 genes in this pathway |
| ROYLANCE BREAST CANCER 16Q COPY NUMBER UP | 63 | 44 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| BENPORATH NANOG TARGETS | 988 | 594 | All SZGR 2.0 genes in this pathway |
| CHEOK RESPONSE TO HD MTX DN | 24 | 18 | All SZGR 2.0 genes in this pathway |
| MUNSHI MULTIPLE MYELOMA UP | 81 | 52 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS LATE PROGENITOR | 544 | 307 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY E2F4 UNSTIMULATED | 728 | 415 | All SZGR 2.0 genes in this pathway |
| HELLER HDAC TARGETS UP | 317 | 208 | All SZGR 2.0 genes in this pathway |
| HELLER HDAC TARGETS SILENCED BY METHYLATION UP | 461 | 298 | All SZGR 2.0 genes in this pathway |
| AMUNDSON POOR SURVIVAL AFTER GAMMA RADIATION 2G | 171 | 96 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER UP | 973 | 570 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER TUMOR VS NORMAL ADJACENT TISSUE UP | 863 | 514 | All SZGR 2.0 genes in this pathway |
| BOCHKIS FOXA2 TARGETS | 425 | 261 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 1HR DN | 222 | 147 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 4HR DN | 254 | 158 | All SZGR 2.0 genes in this pathway |
| KIM BIPOLAR DISORDER OLIGODENDROCYTE DENSITY CORR UP | 682 | 440 | All SZGR 2.0 genes in this pathway |