Gene Page: MRVI1
Summary ?
| GeneID | 10335 |
| Symbol | MRVI1 |
| Synonyms | IRAG|JAW1L |
| Description | murine retrovirus integration site 1 homolog |
| Reference | MIM:604673|HGNC:HGNC:7237|Ensembl:ENSG00000072952|HPRD:05242|Vega:OTTHUMG00000165773 |
| Gene type | protein-coding |
| Map location | 11p15 |
| Pascal p-value | 0.575 |
| Sherlock p-value | 0.872 |
| Fetal beta | -0.588 |
| eGene | Myers' cis & trans Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWAScat | Genome-wide Association Studies | This data set includes 560 SNPs associated with schizophrenia. A total of 486 genes were mapped to these SNPs within 50kb. | |
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs10840431 | chr11 | 10524605 | MRVI1 | 10335 | 4.325E-6 | cis | ||
| rs10770123 | chr11 | 10534865 | MRVI1 | 10335 | 0.08 | cis | ||
| rs10840433 | chr11 | 10547173 | MRVI1 | 10335 | 0.16 | cis | ||
| rs1350991 | chr11 | 10554629 | MRVI1 | 10335 | 0.19 | cis | ||
| rs2054392 | chr11 | 10558598 | MRVI1 | 10335 | 4.089E-6 | cis | ||
| rs10840444 | chr11 | 10624214 | MRVI1 | 10335 | 0.12 | cis | ||
| rs964426 | chr11 | 10632251 | MRVI1 | 10335 | 4.205E-5 | cis | ||
| rs964425 | chr11 | 10632483 | MRVI1 | 10335 | 1.253E-4 | cis | ||
| rs10840431 | chr11 | 10524605 | MRVI1 | 10335 | 7.415E-4 | trans | ||
| rs2054392 | chr11 | 10558598 | MRVI1 | 10335 | 7.061E-4 | trans | ||
| rs964426 | chr11 | 10632251 | MRVI1 | 10335 | 0.01 | trans | ||
| rs964425 | chr11 | 10632483 | MRVI1 | 10335 | 0.01 | trans |
Section II. Transcriptome annotation
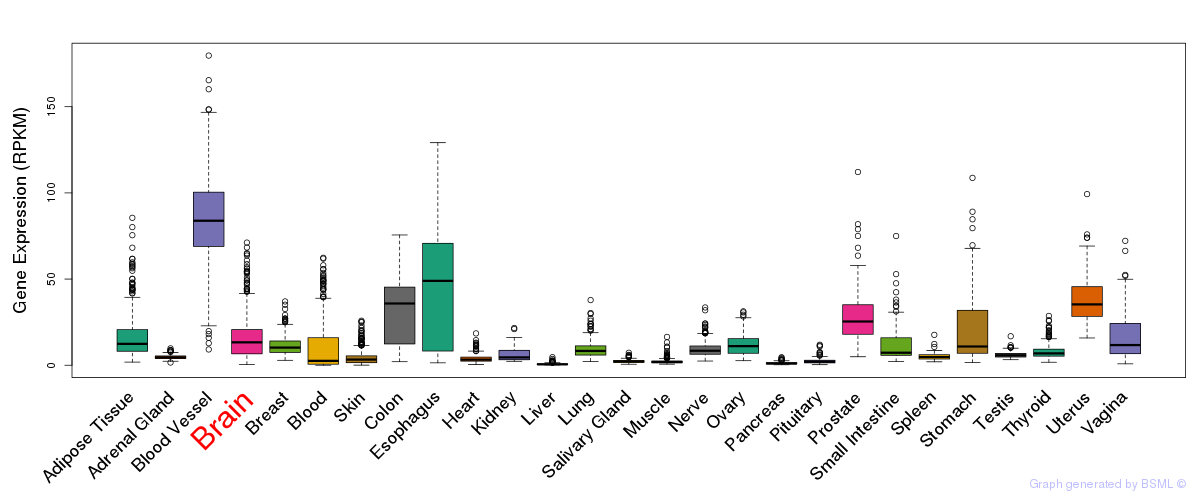
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SMPD4 | 0.94 | 0.94 |
| CREBL1 | 0.94 | 0.93 |
| ZNF384 | 0.94 | 0.94 |
| CNOT3 | 0.94 | 0.93 |
| CIZ1 | 0.94 | 0.94 |
| DHX16 | 0.93 | 0.93 |
| PPIL2 | 0.93 | 0.93 |
| NFRKB | 0.93 | 0.93 |
| DNMT1 | 0.92 | 0.92 |
| GTF3C2 | 0.92 | 0.91 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.31 | -0.77 | -0.84 |
| AF347015.27 | -0.75 | -0.82 |
| MT-CO2 | -0.75 | -0.82 |
| AF347015.21 | -0.72 | -0.89 |
| MT-CYB | -0.72 | -0.79 |
| AF347015.8 | -0.72 | -0.81 |
| AF347015.33 | -0.71 | -0.77 |
| IFI27 | -0.70 | -0.77 |
| HIGD1B | -0.69 | -0.78 |
| C5orf53 | -0.68 | -0.69 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG VASCULAR SMOOTH MUSCLE CONTRACTION | 115 | 81 | All SZGR 2.0 genes in this pathway |
| REACTOME CGMP EFFECTS | 19 | 13 | All SZGR 2.0 genes in this pathway |
| REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | 25 | 19 | All SZGR 2.0 genes in this pathway |
| REACTOME PLATELET HOMEOSTASIS | 78 | 49 | All SZGR 2.0 genes in this pathway |
| REACTOME HEMOSTASIS | 466 | 331 | All SZGR 2.0 genes in this pathway |
| LIU PROSTATE CANCER DN | 481 | 290 | All SZGR 2.0 genes in this pathway |
| BERTUCCI MEDULLARY VS DUCTAL BREAST CANCER DN | 169 | 118 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA POORLY DIFFERENTIATED DN | 805 | 505 | All SZGR 2.0 genes in this pathway |
| CHEN NEUROBLASTOMA COPY NUMBER GAINS | 50 | 35 | All SZGR 2.0 genes in this pathway |
| AFFAR YY1 TARGETS UP | 214 | 133 | All SZGR 2.0 genes in this pathway |
| XU GH1 AUTOCRINE TARGETS UP | 268 | 157 | All SZGR 2.0 genes in this pathway |
| IZADPANAH STEM CELL ADIPOSE VS BONE DN | 108 | 68 | All SZGR 2.0 genes in this pathway |
| CHANDRAN METASTASIS DN | 306 | 191 | All SZGR 2.0 genes in this pathway |
| NIELSEN GIST | 98 | 66 | All SZGR 2.0 genes in this pathway |
| MARTENS BOUND BY PML RARA FUSION | 456 | 287 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS CONFLUENT | 567 | 365 | All SZGR 2.0 genes in this pathway |
| PANGAS TUMOR SUPPRESSION BY SMAD1 AND SMAD5 UP | 134 | 85 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS UP | 504 | 321 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS UP | 745 | 475 | All SZGR 2.0 genes in this pathway |
| HUANG GATA2 TARGETS DN | 72 | 52 | All SZGR 2.0 genes in this pathway |
| PLASARI TGFB1 SIGNALING VIA NFIC 10HR UP | 54 | 38 | All SZGR 2.0 genes in this pathway |
| ACEVEDO FGFR1 TARGETS IN PROSTATE CANCER MODEL DN | 308 | 187 | All SZGR 2.0 genes in this pathway |
| LIM MAMMARY STEM CELL UP | 489 | 314 | All SZGR 2.0 genes in this pathway |