Gene Page: HMG20B
Summary ?
| GeneID | 10362 |
| Symbol | HMG20B |
| Synonyms | BRAF25|BRAF35|HMGX2|HMGXB2|PP7706|SMARCE1r|SOXL|pp8857 |
| Description | high mobility group 20B |
| Reference | MIM:605535|HGNC:HGNC:5002|Ensembl:ENSG00000064961|HPRD:05703|Vega:OTTHUMG00000150437 |
| Gene type | protein-coding |
| Map location | 19p13.3 |
| Pascal p-value | 0.372 |
| DMG | 1 (# studies) |
| Support | Chromatin Remodeling Genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg01335042 | 19 | 3572516 | HMG20B | 8.58E-10 | -0.024 | 1.08E-6 | DMG:Jaffe_2016 |
Section II. Transcriptome annotation
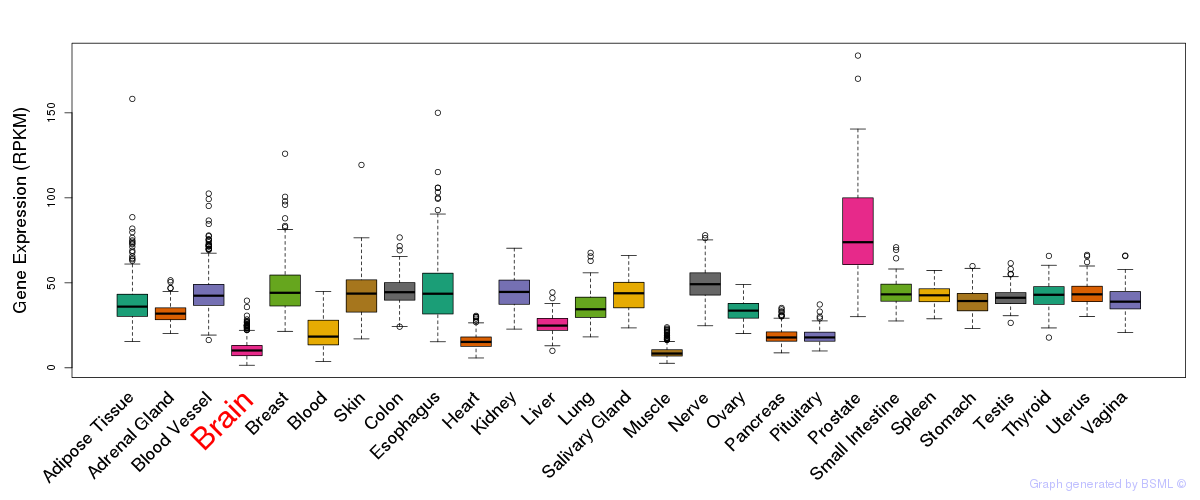
General gene expression (GTEx)
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| RELA | 0.89 | 0.86 |
| JUP | 0.87 | 0.82 |
| WDR6 | 0.86 | 0.79 |
| GLIS2 | 0.86 | 0.89 |
| PLXNB2 | 0.86 | 0.88 |
| ZNF768 | 0.86 | 0.86 |
| CREBL1 | 0.86 | 0.77 |
| MAPK7 | 0.86 | 0.80 |
| AL022328.2 | 0.85 | 0.90 |
| ZNF446 | 0.85 | 0.80 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| C5orf53 | -0.66 | -0.67 |
| ACOT13 | -0.64 | -0.67 |
| PLA2G4A | -0.62 | -0.67 |
| CHN1 | -0.61 | -0.63 |
| FAM162A | -0.61 | -0.65 |
| ACYP2 | -0.61 | -0.64 |
| CA4 | -0.60 | -0.62 |
| CCNI2 | -0.59 | -0.61 |
| CACNA1F | -0.58 | -0.65 |
| TUBA1 | -0.57 | -0.66 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| AOF2 | BHC110 | KDM1 | KIAA0601 | LSD1 | amine oxidase (flavin containing) domain 2 | Affinity Capture-MS Affinity Capture-Western Co-purification | BioGRID | 12032298 |12493763 |
| BRCA2 | BRCC2 | FACD | FAD | FAD1 | FANCB | FANCD | FANCD1 | breast cancer 2, early onset | - | HPRD,BioGRID | 11207365 |
| CCDC85B | DIPA | coiled-coil domain containing 85B | Two-hybrid | BioGRID | 16189514 |
| HDAC1 | DKFZp686H12203 | GON-10 | HD1 | RPD3 | RPD3L1 | histone deacetylase 1 | Affinity Capture-Western Co-purification | BioGRID | 12032298 |
| HDAC2 | RPD3 | YAF1 | histone deacetylase 2 | Affinity Capture-MS Affinity Capture-Western Co-purification | BioGRID | 12032298 |12493763 |
| HNRNPL | FLJ35509 | HNRPL | P/OKcl.14 | hnRNP-L | heterogeneous nuclear ribonucleoprotein L | Two-hybrid | BioGRID | 16169070 |
| KIF4A | FLJ12530 | FLJ12655 | FLJ14204 | FLJ20631 | HSA271784 | KIF4 | KIF4-G1 | kinesin family member 4A | - | HPRD,BioGRID | 12809554 |
| PHF21A | BHC80 | BM-006 | KIAA1696 | PHD finger protein 21A | - | HPRD,BioGRID | 15325272 |
| RCOR1 | COREST | KIAA0071 | RCOR | REST corepressor 1 | Affinity Capture-Western Co-purification | BioGRID | 12032298 |
| SF3A2 | PRP11 | PRPF11 | SAP62 | SF3a66 | splicing factor 3a, subunit 2, 66kDa | Affinity Capture-MS | BioGRID | 12234937 |
| TACC2 | AZU-1 | ECTACC | transforming, acidic coiled-coil containing protein 2 | Two-hybrid | BioGRID | 16169070 |
| VIM | FLJ36605 | vimentin | Two-hybrid | BioGRID | 16169070 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | 132 | 101 | All SZGR 2.0 genes in this pathway |
| REACTOME HEMOSTASIS | 466 | 331 | All SZGR 2.0 genes in this pathway |
| FULCHER INFLAMMATORY RESPONSE LECTIN VS LPS UP | 579 | 346 | All SZGR 2.0 genes in this pathway |
| GARY CD5 TARGETS UP | 473 | 314 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS BASAL UP | 380 | 215 | All SZGR 2.0 genes in this pathway |
| WANG LMO4 TARGETS UP | 372 | 227 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE KERATINOCYTE UP | 530 | 342 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN UP | 1142 | 669 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND DOXORUBICIN UP | 612 | 367 | All SZGR 2.0 genes in this pathway |
| MARTORIATI MDM4 TARGETS FETAL LIVER DN | 514 | 319 | All SZGR 2.0 genes in this pathway |
| DAIRKEE TERT TARGETS UP | 380 | 213 | All SZGR 2.0 genes in this pathway |
| KIM MYC AMPLIFICATION TARGETS DN | 97 | 51 | All SZGR 2.0 genes in this pathway |
| KIM MYCN AMPLIFICATION TARGETS DN | 103 | 59 | All SZGR 2.0 genes in this pathway |
| PUJANA BREAST CANCER LIT INT NETWORK | 101 | 73 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 6HR DN | 514 | 330 | All SZGR 2.0 genes in this pathway |
| BENPORATH CYCLING GENES | 648 | 385 | All SZGR 2.0 genes in this pathway |
| LIN APC TARGETS | 77 | 55 | All SZGR 2.0 genes in this pathway |
| PENG RAPAMYCIN RESPONSE UP | 203 | 130 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| JIANG HYPOXIA CANCER | 83 | 52 | All SZGR 2.0 genes in this pathway |
| HELLER HDAC TARGETS UP | 317 | 208 | All SZGR 2.0 genes in this pathway |
| POS RESPONSE TO HISTAMINE UP | 13 | 10 | All SZGR 2.0 genes in this pathway |
| BLUM RESPONSE TO SALIRASIB DN | 342 | 220 | All SZGR 2.0 genes in this pathway |
| HOSHIDA LIVER CANCER SURVIVAL UP | 73 | 49 | All SZGR 2.0 genes in this pathway |
| WHITFIELD CELL CYCLE G2 M | 216 | 124 | All SZGR 2.0 genes in this pathway |
| WHITFIELD CELL CYCLE M G1 | 148 | 95 | All SZGR 2.0 genes in this pathway |
| VERHAAK GLIOBLASTOMA CLASSICAL | 162 | 122 | All SZGR 2.0 genes in this pathway |
| KASLER HDAC7 TARGETS 1 UP | 194 | 133 | All SZGR 2.0 genes in this pathway |