Gene Page: CACNG2
Summary ?
| GeneID | 10369 |
| Symbol | CACNG2 |
| Synonyms | MRD10 |
| Description | calcium voltage-gated channel auxiliary subunit gamma 2 |
| Reference | MIM:602911|HGNC:HGNC:1406|HPRD:04220| |
| Gene type | protein-coding |
| Map location | 22q13.1 |
| Pascal p-value | 0.104 |
| Sherlock p-value | 0.885 |
| Fetal beta | -1.374 |
| Support | CANABINOID DOPAMINE EXCITABILITY METABOTROPIC GLUTAMATE RECEPTOR SEROTONIN G2Cdb.human_PocklingtonH1 G2Cdb.humanNRC G2Cdb.humanPSD G2Cdb.humanPSP CompositeSet Potential synaptic genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 3 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
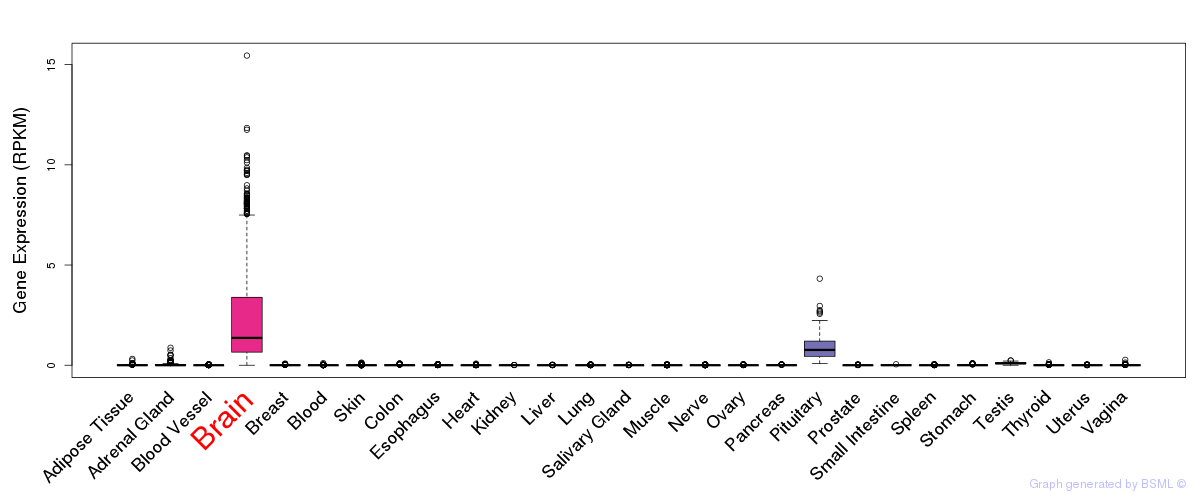
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| PAM | 0.66 | 0.75 |
| GRM8 | 0.66 | 0.71 |
| SPOCK2 | 0.65 | 0.78 |
| PDGFB | 0.65 | 0.68 |
| NHSL2 | 0.62 | 0.68 |
| ARSG | 0.62 | 0.67 |
| B3GALTL | 0.61 | 0.70 |
| PRKCD | 0.61 | 0.67 |
| KCTD8 | 0.61 | 0.68 |
| CCNE1 | 0.61 | 0.64 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| NEUROD6 | -0.44 | -0.33 |
| NEUROD2 | -0.43 | -0.30 |
| KLHL1 | -0.43 | -0.20 |
| RPL34 | -0.41 | -0.55 |
| CSRP2 | -0.41 | -0.45 |
| RPL38 | -0.41 | -0.55 |
| RPL37 | -0.41 | -0.55 |
| PFN3 | -0.40 | -0.42 |
| RPL31 | -0.40 | -0.51 |
| RPL37A | -0.40 | -0.53 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005509 | calcium ion binding | IEA | - | |
| GO:0005244 | voltage-gated ion channel activity | IEA | - | |
| GO:0005245 | voltage-gated calcium channel activity | TAS | 10221464 | |
| GO:0005198 | structural molecule activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007528 | neuromuscular junction development | IEA | Synap (GO term level: 10) | - |
| GO:0019226 | transmission of nerve impulse | IEA | neuron (GO term level: 5) | - |
| GO:0006816 | calcium ion transport | IEA | - | |
| GO:0006811 | ion transport | IEA | - | |
| GO:0042391 | regulation of membrane potential | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005923 | tight junction | IEA | Brain (GO term level: 10) | - |
| GO:0016021 | integral to membrane | IEA | - | |
| GO:0005886 | plasma membrane | EXP | 8825650 | |
| GO:0005891 | voltage-gated calcium channel complex | TAS | 10221464 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG MAPK SIGNALING PATHWAY | 267 | 205 | All SZGR 2.0 genes in this pathway |
| KEGG CARDIAC MUSCLE CONTRACTION | 80 | 51 | All SZGR 2.0 genes in this pathway |
| KEGG HYPERTROPHIC CARDIOMYOPATHY HCM | 85 | 65 | All SZGR 2.0 genes in this pathway |
| KEGG ARRHYTHMOGENIC RIGHT VENTRICULAR CARDIOMYOPATHY ARVC | 76 | 59 | All SZGR 2.0 genes in this pathway |
| KEGG DILATED CARDIOMYOPATHY | 92 | 68 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSMISSION ACROSS CHEMICAL SYNAPSES | 186 | 155 | All SZGR 2.0 genes in this pathway |
| REACTOME NEURONAL SYSTEM | 279 | 221 | All SZGR 2.0 genes in this pathway |
| REACTOME NEUROTRANSMITTER RECEPTOR BINDING AND DOWNSTREAM TRANSMISSION IN THE POSTSYNAPTIC CELL | 137 | 110 | All SZGR 2.0 genes in this pathway |
| REACTOME TRAFFICKING OF AMPA RECEPTORS | 28 | 23 | All SZGR 2.0 genes in this pathway |
| DAIRKEE TERT TARGETS UP | 380 | 213 | All SZGR 2.0 genes in this pathway |
| IWANAGA CARCINOGENESIS BY KRAS PTEN DN | 353 | 226 | All SZGR 2.0 genes in this pathway |
| MEISSNER NPC HCP WITH H3 UNMETHYLATED | 536 | 296 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN IPS WITH HCP H3K27ME3 | 102 | 76 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN ES HCP WITH H3 UNMETHYLATED | 63 | 31 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MEF HCP WITH H3 UNMETHYLATED | 228 | 119 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP A | 898 | 516 | All SZGR 2.0 genes in this pathway |
| ZWANG CLASS 1 TRANSIENTLY INDUCED BY EGF | 516 | 308 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-129-5p | 158 | 164 | 1A | hsa-miR-129brain | CUUUUUGCGGUCUGGGCUUGC |
| hsa-miR-129-5p | CUUUUUGCGGUCUGGGCUUGCU | ||||
| miR-375 | 200 | 206 | 1A | hsa-miR-375 | UUUGUUCGUUCGGCUCGCGUGA |
| miR-450 | 102 | 108 | 1A | hsa-miR-450 | UUUUUGCGAUGUGUUCCUAAUA |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.