Gene Page: TESK2
Summary ?
| GeneID | 10420 |
| Symbol | TESK2 |
| Synonyms | - |
| Description | testis-specific kinase 2 |
| Reference | MIM:604746|HGNC:HGNC:11732|Ensembl:ENSG00000070759|Vega:OTTHUMG00000007680 |
| Gene type | protein-coding |
| Map location | 1p32 |
| Pascal p-value | 0.84 |
| Sherlock p-value | 0.559 |
| Fetal beta | -0.362 |
| DMG | 1 (# studies) |
| eGene | Caudate basal ganglia Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg25827112 | 1 | 45956773 | TESK2 | 1.55E-8 | -0.008 | 5.83E-6 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs6684566 | chr1 | 238223494 | TESK2 | 10420 | 0.18 | trans | ||
| rs5942141 | chrX | 91433860 | TESK2 | 10420 | 0.19 | trans |
Section II. Transcriptome annotation
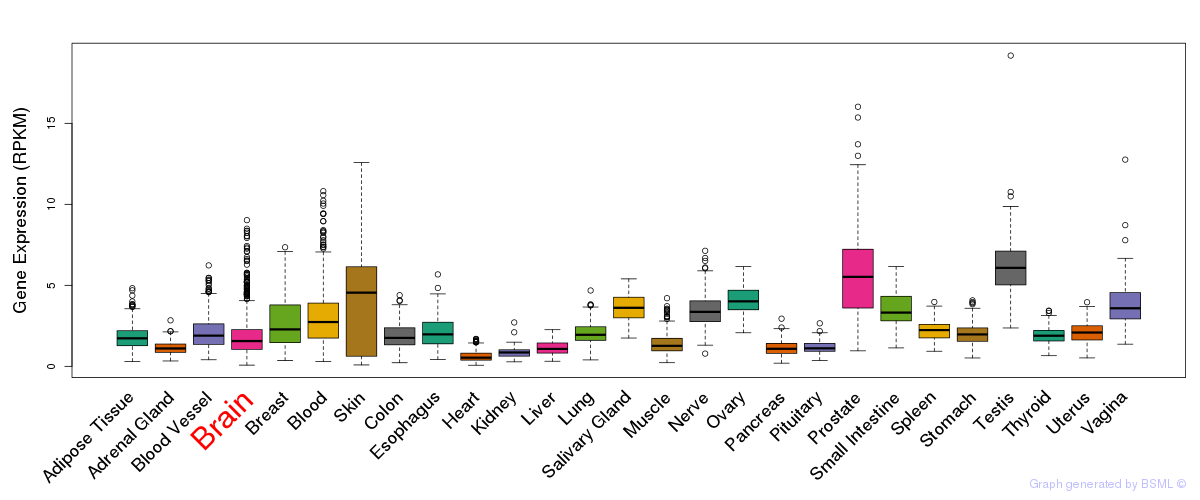
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MRPS24 | 0.90 | 0.89 |
| NDUFA11 | 0.89 | 0.89 |
| MIF | 0.89 | 0.88 |
| NUDT16L1 | 0.89 | 0.89 |
| SCAND1 | 0.88 | 0.86 |
| DGCR6L | 0.87 | 0.85 |
| MRPL12 | 0.87 | 0.85 |
| ARF5 | 0.87 | 0.86 |
| PRDX2 | 0.86 | 0.84 |
| CCDC56 | 0.86 | 0.83 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AP000769.2 | -0.47 | -0.61 |
| Z83840.4 | -0.44 | -0.47 |
| AF347015.26 | -0.43 | -0.44 |
| MALAT1 | -0.43 | -0.53 |
| AC016705.1 | -0.41 | -0.47 |
| AF347015.18 | -0.40 | -0.42 |
| CCDC55 | -0.39 | -0.38 |
| AC008088.2 | -0.39 | -0.49 |
| AF347015.2 | -0.39 | -0.39 |
| COBLL1 | -0.39 | -0.37 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| ENK UV RESPONSE EPIDERMIS DN | 508 | 354 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 3 UP | 329 | 196 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC MAX TARGETS | 775 | 494 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 16HR UP | 225 | 139 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 12HR UP | 111 | 68 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 STIMULATED | 1022 | 619 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 UNSTIMULATED | 1229 | 713 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY E2F4 UNSTIMULATED | 728 | 415 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| PURBEY TARGETS OF CTBP1 NOT SATB1 DN | 448 | 282 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 1ST EGF PULSE ONLY | 1839 | 928 | All SZGR 2.0 genes in this pathway |
| ZWANG EGF INTERVAL DN | 214 | 124 | All SZGR 2.0 genes in this pathway |