Gene Page: UBAC1
Summary ?
| GeneID | 10422 |
| Symbol | UBAC1 |
| Synonyms | GBDR1|UBADC1 |
| Description | UBA domain containing 1 |
| Reference | MIM:608129|HGNC:HGNC:30221|Ensembl:ENSG00000130560|HPRD:12170|Vega:OTTHUMG00000020920 |
| Gene type | protein-coding |
| Map location | 9q34.3 |
| Pascal p-value | 0.638 |
| Sherlock p-value | 0.257 |
| Fetal beta | -0.214 |
| eGene | Cerebellar Hemisphere Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
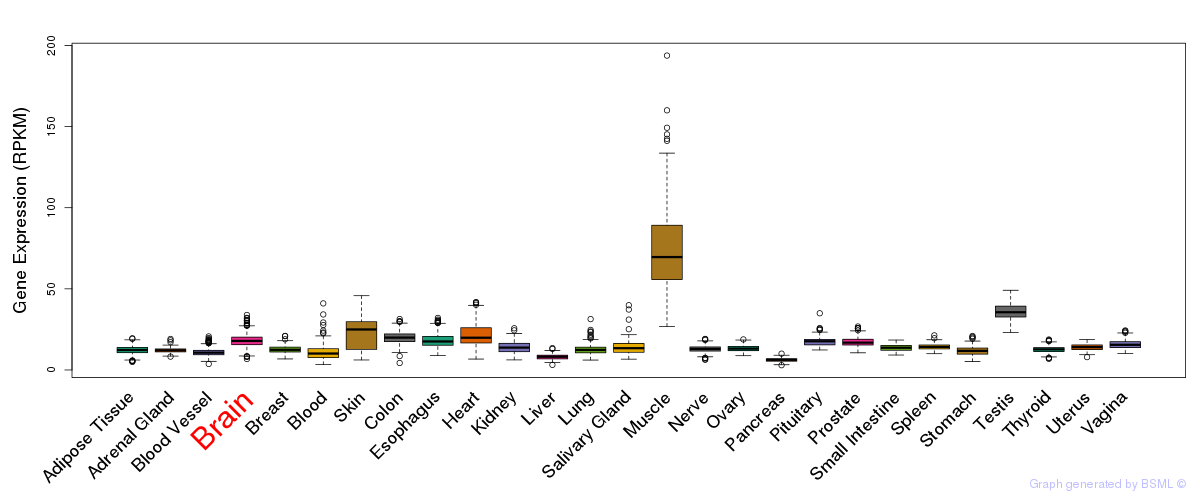
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| USO1 | 0.88 | 0.88 |
| EIF5 | 0.88 | 0.89 |
| HBS1L | 0.87 | 0.89 |
| AGGF1 | 0.85 | 0.86 |
| NARS | 0.85 | 0.86 |
| DLD | 0.85 | 0.88 |
| SLC4A1AP | 0.84 | 0.87 |
| ARMCX3 | 0.84 | 0.85 |
| UCHL5 | 0.84 | 0.84 |
| FMR1 | 0.84 | 0.85 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.31 | -0.58 | -0.55 |
| AF347015.2 | -0.58 | -0.51 |
| CST3 | -0.58 | -0.58 |
| AF347015.21 | -0.58 | -0.50 |
| MT-CO2 | -0.58 | -0.53 |
| HIGD1B | -0.57 | -0.56 |
| GATSL3 | -0.57 | -0.61 |
| METRN | -0.57 | -0.58 |
| ENHO | -0.56 | -0.67 |
| APOE | -0.56 | -0.56 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| TIEN INTESTINE PROBIOTICS 24HR UP | 557 | 331 | All SZGR 2.0 genes in this pathway |
| LOPEZ MBD TARGETS | 957 | 597 | All SZGR 2.0 genes in this pathway |
| CAFFAREL RESPONSE TO THC DN | 31 | 23 | All SZGR 2.0 genes in this pathway |
| STARK PREFRONTAL CORTEX 22Q11 DELETION DN | 517 | 309 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS MATURE CELL | 293 | 160 | All SZGR 2.0 genes in this pathway |
| KAAB HEART ATRIUM VS VENTRICLE DN | 261 | 183 | All SZGR 2.0 genes in this pathway |
| RAMASWAMY METASTASIS UP | 66 | 43 | All SZGR 2.0 genes in this pathway |
| STEIN ESRRA TARGETS UP | 388 | 234 | All SZGR 2.0 genes in this pathway |
| FUJII YBX1 TARGETS DN | 202 | 132 | All SZGR 2.0 genes in this pathway |
| ACEVEDO NORMAL TISSUE ADJACENT TO LIVER TUMOR DN | 354 | 216 | All SZGR 2.0 genes in this pathway |
| SHEDDEN LUNG CANCER POOR SURVIVAL A6 | 456 | 285 | All SZGR 2.0 genes in this pathway |
| STEIN ESRRA TARGETS | 535 | 325 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| CHYLA CBFA2T3 TARGETS DN | 242 | 146 | All SZGR 2.0 genes in this pathway |
| WIERENGA STAT5A TARGETS UP | 217 | 131 | All SZGR 2.0 genes in this pathway |
| WIERENGA STAT5A TARGETS GROUP1 | 136 | 76 | All SZGR 2.0 genes in this pathway |
| LINSLEY MIR16 TARGETS | 206 | 127 | All SZGR 2.0 genes in this pathway |