Gene Page: MCRS1
Summary ?
| GeneID | 10445 |
| Symbol | MCRS1 |
| Synonyms | ICP22BP|INO80Q|MCRS2|MSP58|P78 |
| Description | microspherule protein 1 |
| Reference | MIM:609504|HGNC:HGNC:6960|Ensembl:ENSG00000187778|HPRD:11298|Vega:OTTHUMG00000169620 |
| Gene type | protein-coding |
| Map location | 12q13.12 |
| Pascal p-value | 0.027 |
| Sherlock p-value | 0.598 |
| Fetal beta | -0.359 |
| DMG | 1 (# studies) |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg00699235 | 12 | 49962218 | MCRS1 | 7.35E-10 | -0.021 | 9.96E-7 | DMG:Jaffe_2016 |
Section II. Transcriptome annotation
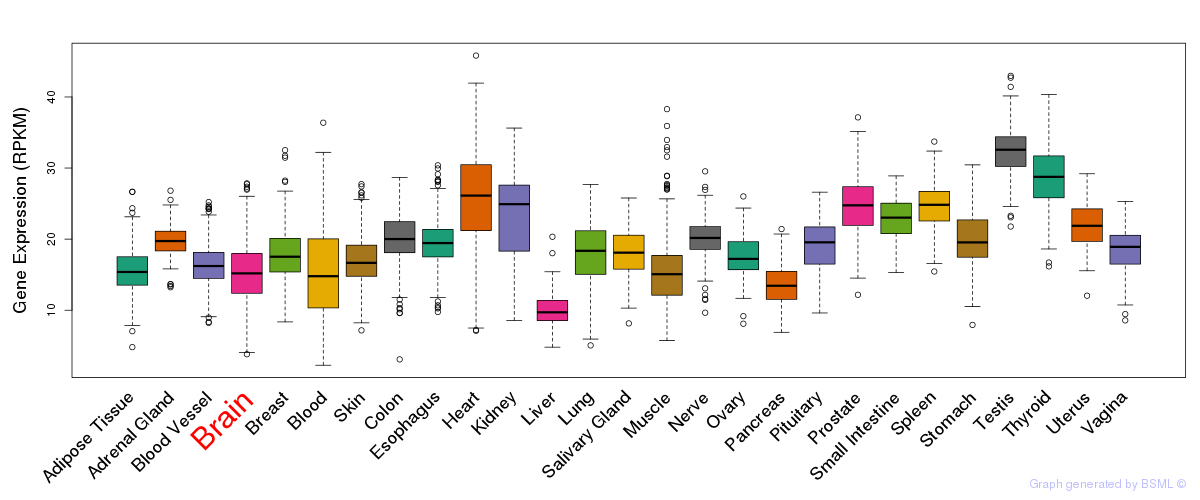
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| C14orf106 | 0.93 | 0.89 |
| CKAP2 | 0.90 | 0.82 |
| MASTL | 0.90 | 0.86 |
| ATAD2 | 0.89 | 0.84 |
| ECT2 | 0.88 | 0.82 |
| SMARCA5 | 0.87 | 0.87 |
| CENPC1 | 0.87 | 0.84 |
| SGOL2 | 0.87 | 0.82 |
| SMC4 | 0.87 | 0.79 |
| TMEM194A | 0.87 | 0.84 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.31 | -0.56 | -0.73 |
| MT-CO2 | -0.55 | -0.73 |
| FXYD1 | -0.54 | -0.70 |
| AF347015.33 | -0.54 | -0.69 |
| MT-CYB | -0.53 | -0.69 |
| AF347015.27 | -0.53 | -0.69 |
| AF347015.8 | -0.53 | -0.71 |
| HIGD1B | -0.52 | -0.69 |
| AF347015.21 | -0.52 | -0.72 |
| IFI27 | -0.52 | -0.68 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| C19orf50 | FLJ25480 | MGC2749 | MST096 | MSTP096 | chromosome 19 open reading frame 50 | Two-hybrid | BioGRID | 16189514 |
| CCDC53 | CGI-116 | coiled-coil domain containing 53 | Two-hybrid | BioGRID | 16189514 |
| CCDC85B | DIPA | coiled-coil domain containing 85B | Two-hybrid | BioGRID | 16189514 |
| CSNK2B | CK2B | CK2N | CSK2B | G5A | MGC138222 | MGC138224 | casein kinase 2, beta polypeptide | Affinity Capture-MS | BioGRID | 17353931 |
| DAXX | BING2 | DAP6 | EAP1 | MGC126245 | MGC126246 | death-domain associated protein | - | HPRD,BioGRID | 11948183 |
| DAXX | BING2 | DAP6 | EAP1 | MGC126245 | MGC126246 | death-domain associated protein | Daxx interacts with MCRS1 (MSP58). | BIND | 11948183 |
| DVL2 | - | dishevelled, dsh homolog 2 (Drosophila) | Two-hybrid | BioGRID | 16189514 |
| FBL | FIB | FLRN | RNU3IP1 | fibrillarin | Co-fractionation | BioGRID | 9654073 |
| FXR2 | FMR1L2 | fragile X mental retardation, autosomal homolog 2 | Two-hybrid | BioGRID | 16189514 |
| KIAA1712 | PS1TP3 | KIAA1712 | Two-hybrid | BioGRID | 16189514 |
| LSM6 | YDR378C | LSM6 homolog, U6 small nuclear RNA associated (S. cerevisiae) | Two-hybrid | BioGRID | 15231747 |
| MAGEA11 | MAGE-11 | MAGE11 | MAGEA-11 | MGC10511 | melanoma antigen family A, 11 | Two-hybrid | BioGRID | 16189514 |
| MED4 | DRIP36 | FLJ10956 | HSPC126 | RP11-90M2.2 | TRAP36 | VDRIP | mediator complex subunit 4 | Two-hybrid | BioGRID | 16189514 |
| NOP2 | MGC117384 | MGC149287 | MGC149288 | NOL1 | NOP120 | NSUN1 | p120 | NOP2 nucleolar protein homolog (yeast) | - | HPRD,BioGRID | 9654073 |
| PBX2 | G17 | HOX12 | PBX2MHC | pre-B-cell leukemia homeobox 2 | - | HPRD | 14667819 |
| PHC2 | EDR2 | HPH2 | MGC163502 | PH2 | polyhomeotic homolog 2 (Drosophila) | Affinity Capture-Western Two-hybrid | BioGRID | 16189514 |
| PINX1 | FLJ20565 | LPTL | LPTS | MGC8850 | PIN2-interacting protein 1 | - | HPRD,BioGRID | 15044100 |
| PRKAR1A | CAR | CNC | CNC1 | DKFZp779L0468 | MGC17251 | PKR1 | PPNAD1 | PRKAR1 | TSE1 | protein kinase, cAMP-dependent, regulatory, type I, alpha (tissue specific extinguisher 1) | Affinity Capture-MS | BioGRID | 17353931 |
| RALYL | HNRPCL3 | RALY RNA binding protein-like | Two-hybrid | BioGRID | 16189514 |
| TERT | EST2 | TCS1 | TP2 | TRT | hEST2 | telomerase reverse transcriptase | - | HPRD,BioGRID | 15044100 |
| TRIM37 | KIAA0898 | MUL | POB1 | TEF3 | tripartite motif-containing 37 | Two-hybrid | BioGRID | 16189514 |
| UBR4 | FLJ41863 | KIAA0462 | KIAA1307 | RBAF600 | ZUBR1 | p600 | ubiquitin protein ligase E3 component n-recognin 4 | Affinity Capture-MS | BioGRID | 17353931 |
| UBR5 | DD5 | EDD | EDD1 | FLJ11310 | HYD | KIAA0896 | MGC57263 | ubiquitin protein ligase E3 component n-recognin 5 | Affinity Capture-MS | BioGRID | 17353931 |
| UPF3B | HUPF3B | MRXS14 | RENT3B | UPF3X | UPF3 regulator of nonsense transcripts homolog B (yeast) | - | HPRD,BioGRID | 15231747 |
| WBP11 | DKFZp779M1063 | NPWBP | SIPP1 | WW domain binding protein 11 | Two-hybrid | BioGRID | 16189514 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| PATIL LIVER CANCER | 747 | 453 | All SZGR 2.0 genes in this pathway |
| HASLINGER B CLL WITH CHROMOSOME 12 TRISOMY | 24 | 12 | All SZGR 2.0 genes in this pathway |
| ICHIBA GRAFT VERSUS HOST DISEASE 35D UP | 131 | 79 | All SZGR 2.0 genes in this pathway |
| AGUIRRE PANCREATIC CANCER COPY NUMBER DN | 238 | 145 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |