Gene Page: CDX2
Summary ?
| GeneID | 1045 |
| Symbol | CDX2 |
| Synonyms | CDX-3|CDX2/AS|CDX3 |
| Description | caudal type homeobox 2 |
| Reference | MIM:600297|HGNC:HGNC:1806|HPRD:02622| |
| Gene type | protein-coding |
| Map location | 13q12.3 |
| Pascal p-value | 0.936 |
| Fetal beta | -0.413 |
| DMG | 1 (# studies) |
| eGene | Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg02550231 | 13 | 28542287 | CDX2 | 7.96E-7 | -0.923 | 0.007 | DMG:Wockner_2014 |
Section II. Transcriptome annotation
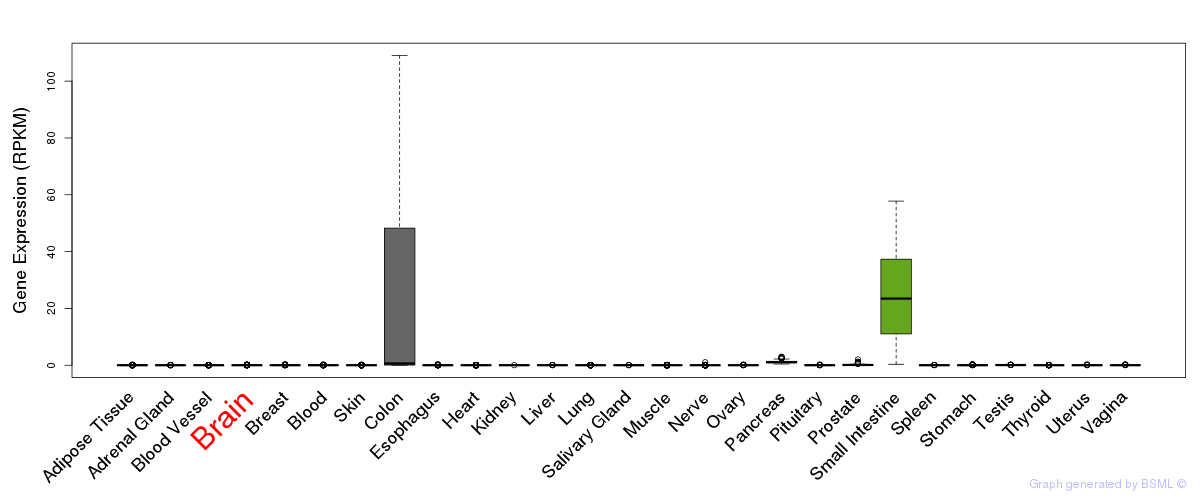
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MT2A | 0.80 | 0.70 |
| SRGN | 0.79 | 0.79 |
| IFITM2 | 0.78 | 0.76 |
| C13orf15 | 0.77 | 0.77 |
| C10orf54 | 0.76 | 0.79 |
| MT1A | 0.75 | 0.67 |
| GADD45B | 0.74 | 0.82 |
| IFITM3 | 0.74 | 0.74 |
| MT1P2 | 0.73 | 0.66 |
| ATOH8 | 0.73 | 0.74 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AGPAT6 | -0.53 | -0.59 |
| FHOD3 | -0.51 | -0.57 |
| ING2 | -0.51 | -0.57 |
| RTF1 | -0.51 | -0.61 |
| IKBKAP | -0.51 | -0.56 |
| RNASEN | -0.51 | -0.59 |
| USP7 | -0.51 | -0.50 |
| TP53BP1 | -0.51 | -0.54 |
| IWS1 | -0.51 | -0.57 |
| SUPV3L1 | -0.51 | -0.60 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| REACTOME INTEGRATION OF ENERGY METABOLISM | 120 | 84 | All SZGR 2.0 genes in this pathway |
| REACTOME REGULATION OF INSULIN SECRETION | 93 | 65 | All SZGR 2.0 genes in this pathway |
| REACTOME INCRETIN SYNTHESIS SECRETION AND INACTIVATION | 22 | 14 | All SZGR 2.0 genes in this pathway |
| REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | 19 | 12 | All SZGR 2.0 genes in this pathway |
| RODRIGUES NTN1 TARGETS DN | 158 | 102 | All SZGR 2.0 genes in this pathway |
| SCHLESINGER METHYLATED IN COLON CANCER | 10 | 7 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| SCHLESINGER METHYLATED DE NOVO IN CANCER | 88 | 64 | All SZGR 2.0 genes in this pathway |
| BENPORATH SUZ12 TARGETS | 1038 | 678 | All SZGR 2.0 genes in this pathway |
| BENPORATH EED TARGETS | 1062 | 725 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES WITH H3K27ME3 | 1118 | 744 | All SZGR 2.0 genes in this pathway |
| BENPORATH PRC2 TARGETS | 652 | 441 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL QTL TRANS | 882 | 572 | All SZGR 2.0 genes in this pathway |
| MAHAJAN RESPONSE TO IL1A UP | 81 | 52 | All SZGR 2.0 genes in this pathway |
| SANSOM APC MYC TARGETS | 217 | 138 | All SZGR 2.0 genes in this pathway |
| HOQUE METHYLATED IN CANCER | 56 | 45 | All SZGR 2.0 genes in this pathway |
| GRESHOCK CANCER COPY NUMBER UP | 323 | 240 | All SZGR 2.0 genes in this pathway |
| MEISSNER NPC HCP WITH H3K27ME3 | 79 | 59 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K27ME3 | 269 | 159 | All SZGR 2.0 genes in this pathway |
| JISON SICKLE CELL DISEASE DN | 181 | 97 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN NPC HCP WITH H3K27ME3 | 341 | 243 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MEF HCP WITH H3K27ME3 | 590 | 403 | All SZGR 2.0 genes in this pathway |
| KIM BIPOLAR DISORDER OLIGODENDROCYTE DENSITY CORR DN | 88 | 59 | All SZGR 2.0 genes in this pathway |
| PURBEY TARGETS OF CTBP1 AND SATB1 UP | 87 | 52 | All SZGR 2.0 genes in this pathway |
| JUBAN TARGETS OF SPI1 AND FLI1 UP | 115 | 73 | All SZGR 2.0 genes in this pathway |
| KOHOUTEK CCNT2 TARGETS | 58 | 35 | All SZGR 2.0 genes in this pathway |