Gene Page: KAT5
Summary ?
| GeneID | 10524 |
| Symbol | KAT5 |
| Synonyms | ESA1|HTATIP|HTATIP1|PLIP|TIP|TIP60|ZC2HC5|cPLA2 |
| Description | lysine acetyltransferase 5 |
| Reference | MIM:601409|HGNC:HGNC:5275|Ensembl:ENSG00000172977|HPRD:03245|Vega:OTTHUMG00000166617 |
| Gene type | protein-coding |
| Map location | 11q13 |
| Pascal p-value | 0.006 |
| DEG p-value | DEG:Maycox_2009:CC_BA10_fold_change=-1.05:CC_BA10_disease_P=0.0415:HBB_BA9_fold_change=-1.12:HBB_BA9_disease_P=0.0352 |
| Fetal beta | 0.164 |
| Support | Chromatin Remodeling Genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DEG:Maycox_2009 | Microarray to determine the expression of over 30000 mRNA transcripts in post-mortem tissue | We included 51 genes whose expression changes are common between two schizophrenia cohorts. | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.8537 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
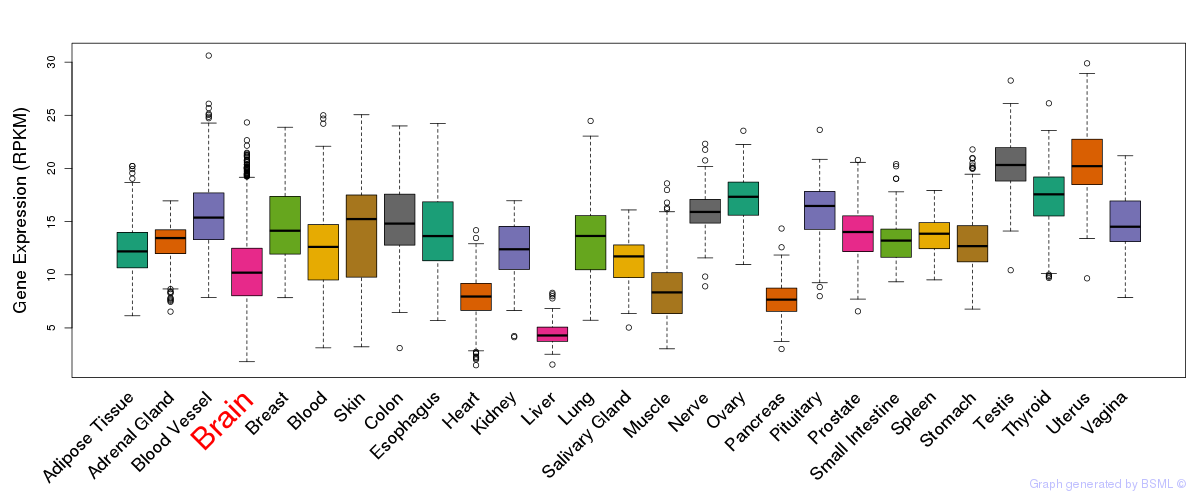
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| VBP1 | 0.89 | 0.87 |
| PSMD10 | 0.86 | 0.86 |
| PSMD14 | 0.86 | 0.85 |
| FH | 0.86 | 0.84 |
| GTF2B | 0.85 | 0.85 |
| TCEAL8 | 0.85 | 0.82 |
| YWHAE | 0.85 | 0.85 |
| EAPP | 0.85 | 0.86 |
| DRG1 | 0.85 | 0.84 |
| MORF4L1 | 0.85 | 0.87 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MT-CO2 | -0.61 | -0.57 |
| AF347015.8 | -0.60 | -0.56 |
| AF347015.2 | -0.60 | -0.60 |
| AF347015.26 | -0.59 | -0.59 |
| AF347015.33 | -0.59 | -0.57 |
| MT-CYB | -0.58 | -0.56 |
| TINAGL1 | -0.58 | -0.62 |
| IFI27 | -0.57 | -0.62 |
| USHBP1 | -0.57 | -0.62 |
| AF347015.15 | -0.57 | -0.57 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003682 | chromatin binding | IEA | - | |
| GO:0003713 | transcription coactivator activity | NAS | 15572661 | |
| GO:0005515 | protein binding | IPI | 15383276 |16169070 |17110330 | |
| GO:0004402 | histone acetyltransferase activity | IEA | - | |
| GO:0016740 | transferase activity | IEA | - | |
| GO:0008270 | zinc ion binding | IEA | - | |
| GO:0008415 | acyltransferase activity | IEA | - | |
| GO:0046872 | metal ion binding | IEA | - | |
| GO:0050681 | androgen receptor binding | NAS | 15572661 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0001558 | regulation of cell growth | IEA | - | |
| GO:0006302 | double-strand break repair | IMP | 10966108 | |
| GO:0006333 | chromatin assembly or disassembly | IEA | - | |
| GO:0006350 | transcription | IEA | - | |
| GO:0006366 | transcription from RNA polymerase II promoter | TAS | 8607265 | |
| GO:0016568 | chromatin modification | IEA | - | |
| GO:0016573 | histone acetylation | IDA | 10966108 | |
| GO:0030521 | androgen receptor signaling pathway | NAS | 15572661 | |
| GO:0045893 | positive regulation of transcription, DNA-dependent | NAS | 15572661 | |
| GO:0044419 | interspecies interaction between organisms | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0000785 | chromatin | IEA | - | |
| GO:0005634 | nucleus | TAS | 8607265 | |
| GO:0005737 | cytoplasm | IEA | - | |
| GO:0035267 | NuA4 histone acetyltransferase complex | IDA | 10966108 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ACTL6A | ACTL6 | Arp4 | BAF53A | INO80K | MGC5382 | actin-like 6A | Co-fractionation | BioGRID | 11839798 |
| APBB1 | FE65 | MGC:9072 | RIR | amyloid beta (A4) precursor protein-binding, family B, member 1 (Fe65) | - | HPRD | 12888553 |
| APP | AAA | ABETA | ABPP | AD1 | APPI | CTFgamma | CVAP | PN2 | amyloid beta (A4) precursor protein | - | HPRD | 12888553 |
| AR | AIS | DHTR | HUMARA | KD | NR3C4 | SBMA | SMAX1 | TFM | androgen receptor | Biochemical Activity Two-hybrid | BioGRID | 11994312 |
| AR | AIS | DHTR | HUMARA | KD | NR3C4 | SBMA | SMAX1 | TFM | androgen receptor | - | HPRD | 10364196 |
| ARIH2 | ARI2 | FLJ10938 | FLJ33921 | TRIAD1 | ariadne homolog 2 (Drosophila) | Two-hybrid | BioGRID | 16169070 |
| BCL3 | BCL4 | D19S37 | B-cell CLL/lymphoma 3 | - | HPRD,BioGRID | 10362352 |
| BCL3 | BCL4 | D19S37 | B-cell CLL/lymphoma 3 | HTATIP (Tip60) interacts with BCL3. | BIND | 15829968 |
| BMI1 | MGC12685 | PCGF4 | RNF51 | BMI1 polycomb ring finger oncogene | Two-hybrid | BioGRID | 16169070 |
| C1orf174 | RP13-531C17.2 | chromosome 1 open reading frame 174 | Two-hybrid | BioGRID | 16169070 |
| C20orf20 | Eaf7 | FLJ10914 | MRG15BP | MRGBP | URCC4 | chromosome 20 open reading frame 20 | - | HPRD | 12963728 |
| C7orf20 | CEE | CGI-20 | chromosome 7 open reading frame 20 | Two-hybrid | BioGRID | 16169070 |
| CBX8 | HPC3 | PC3 | RC1 | chromobox homolog 8 (Pc class homolog, Drosophila) | Two-hybrid | BioGRID | 16169070 |
| CCDC106 | HSU79303 | ZNF581 | coiled-coil domain containing 106 | Two-hybrid | BioGRID | 16169070 |
| CCT7 | CCT-ETA | Ccth | MGC110985 | Nip7-1 | TCP-1-eta | chaperonin containing TCP1, subunit 7 (eta) | Two-hybrid | BioGRID | 16169070 |
| CD82 | 4F9 | C33 | GR15 | IA4 | KAI1 | R2 | SAR2 | ST6 | TSPAN27 | CD82 molecule | HTATIP (Tip60) interacts with the CD82 (KAI1) promoter. | BIND | 15829968 |
| CDC42 | CDC42Hs | G25K | cell division cycle 42 (GTP binding protein, 25kDa) | Two-hybrid | BioGRID | 16169070 |
| CDK5RAP2 | C48 | Cep215 | DKFZp686B1070 | DKFZp686D1070 | KIAA1633 | MCPH3 | CDK5 regulatory subunit associated protein 2 | Two-hybrid | BioGRID | 16169070 |
| CREB1 | CREB | MGC9284 | cAMP responsive element binding protein 1 | - | HPRD,BioGRID | 10720489 |
| CRELD1 | AVSD2 | CIRRIN | DKFZp566D213 | cysteine-rich with EGF-like domains 1 | Two-hybrid | BioGRID | 16169070 |
| CSTF2 | CstF-64 | cleavage stimulation factor, 3' pre-RNA, subunit 2, 64kDa | Two-hybrid | BioGRID | 16169070 |
| DLEU1 | BCMS | DLB1 | DLEU2 | LEU1 | LEU2 | MGC22430 | NCRNA00021 | XTP6 | deleted in lymphocytic leukemia 1 (non-protein coding) | Two-hybrid | BioGRID | 16169070 |
| EDNRA | ETA | ETRA | endothelin receptor type A | Affinity Capture-Western Reconstituted Complex Two-hybrid | BioGRID | 11262386 |
| EFNA1 | B61 | ECKLG | EFL1 | EPLG1 | LERK1 | TNFAIP4 | ephrin-A1 | Two-hybrid | BioGRID | 16169070 |
| EP400 | CAGH32 | DKFZP434I225 | FLJ42018 | FLJ45115 | P400 | TNRC12 | E1A binding protein p400 | Affinity Capture-Western | BioGRID | 11509179 |
| ETV6 | TEL | TEL/ABL | ets variant 6 | - | HPRD,BioGRID | 12737628 |
| FAM101B | MGC45871 | family with sequence similarity 101, member B | Two-hybrid | BioGRID | 16169070 |
| FAM135B | C8ORFK32 | MGC126009 | MGC126010 | MGC33221 | family with sequence similarity 135, member B | Two-hybrid | BioGRID | 16169070 |
| FAM173A | C16orf24 | MGC2494 | family with sequence similarity 173, member A | Two-hybrid | BioGRID | 16169070 |
| FAS | ALPS1A | APO-1 | APT1 | CD95 | FAS1 | FASTM | TNFRSF6 | Fas (TNF receptor superfamily, member 6) | HTATIP (Tip60) interacts with the FAS promoter. | BIND | 15829968 |
| GEMIN7 | SIP3 | gem (nuclear organelle) associated protein 7 | Two-hybrid | BioGRID | 16169070 |
| GRIN1 | NMDA1 | NMDAR1 | NR1 | glutamate receptor, ionotropic, N-methyl D-aspartate 1 | - | HPRD | 10862698 |
| GSTO1 | DKFZp686H13163 | GSTTLp28 | P28 | glutathione S-transferase omega 1 | Two-hybrid | BioGRID | 16169070 |
| HDAC1 | DKFZp686H12203 | GON-10 | HD1 | RPD3 | RPD3L1 | histone deacetylase 1 | - | HPRD | 11994312 |
| HDAC7 | DKFZp586J0917 | FLJ99588 | HD7A | HDAC7A | histone deacetylase 7 | Affinity Capture-Western Two-hybrid | BioGRID | 12551922 |
| HNRNPH3 | 2H9 | FLJ34092 | HNRPH3 | heterogeneous nuclear ribonucleoprotein H3 (2H9) | Two-hybrid | BioGRID | 16169070 |
| IK | CSA2 | MGC59741 | RED | IK cytokine, down-regulator of HLA II | Two-hybrid | BioGRID | 16169070 |
| KAT5 | ESA1 | HTATIP | HTATIP1 | PLIP | TIP | TIP60 | cPLA2 | K(lysine) acetyltransferase 5 | Affinity Capture-Western | BioGRID | 11416127 |
| KLK3 | APS | KLK2A1 | PSA | hK3 | kallikrein-related peptidase 3 | HTATIP (Tip60) interacts with the KLK3 (PSA) promoter. | BIND | 15829968 |
| LMNA | CDCD1 | CDDC | CMD1A | CMT2B1 | EMD2 | FPL | FPLD | HGPS | IDC | LDP1 | LFP | LGMD1B | LMN1 | LMNC | PRO1 | lamin A/C | Two-hybrid | BioGRID | 16169070 |
| LRP1 | A2MR | APOER | APR | CD91 | FLJ16451 | IGFBP3R | LRP | MGC88725 | TGFBR5 | low density lipoprotein-related protein 1 (alpha-2-macroglobulin receptor) | - | HPRD,BioGRID | 12888553 |
| MAD2L1BP | CMT2 | KIAA0110 | MGC11282 | RP1-261G23.6 | MAD2L1 binding protein | Two-hybrid | BioGRID | 16169070 |
| MDM2 | HDMX | MGC71221 | hdm2 | Mdm2 p53 binding protein homolog (mouse) | - | HPRD,BioGRID | 11927554 |
| MYC | bHLHe39 | c-Myc | v-myc myelocytomatosis viral oncogene homolog (avian) | - | HPRD,BioGRID | 12776177 |
| NAP1L5 | DRLM | nucleosome assembly protein 1-like 5 | Two-hybrid | BioGRID | 16169070 |
| NDUFA4L2 | FLJ26118 | NUOMS | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4-like 2 | Two-hybrid | BioGRID | 16169070 |
| NDUFV2 | - | NADH dehydrogenase (ubiquinone) flavoprotein 2, 24kDa | Two-hybrid | BioGRID | 16169070 |
| NFKB1 | DKFZp686C01211 | EBP-1 | KBF1 | MGC54151 | NF-kappa-B | NFKB-p105 | NFKB-p50 | p105 | nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 | HTATIP (Tip60) interacts with NFKB1 (p50). | BIND | 15829968 |
| OGFOD2 | DKFZp686H15154 | FLJ13491 | FLJ37501 | MGC120434 | MGC120436 | 2-oxoglutarate and iron-dependent oxygenase domain containing 2 | Two-hybrid | BioGRID | 16169070 |
| PDCD5 | MGC9294 | TFAR19 | programmed cell death 5 | Two-hybrid | BioGRID | 16169070 |
| PLA2G4A | MGC126350 | PLA2G4 | cPLA2-alpha | phospholipase A2, group IVA (cytosolic, calcium-dependent) | - | HPRD,BioGRID | 11416127 |
| PLEKHA4 | PEPP1 | pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 4 | Two-hybrid | BioGRID | 16169070 |
| POLE2 | DPE2 | polymerase (DNA directed), epsilon 2 (p59 subunit) | Two-hybrid | BioGRID | 16169070 |
| POLR3F | MGC13517 | RPC39 | RPC6 | polymerase (RNA) III (DNA directed) polypeptide F, 39 kDa | Two-hybrid | BioGRID | 16169070 |
| PTPN4 | PTPMEG | PTPMEG1 | protein tyrosine phosphatase, non-receptor type 4 (megakaryocyte) | Two-hybrid | BioGRID | 16169070 |
| PTPRS | PTPSIGMA | protein tyrosine phosphatase, receptor type, S | Two-hybrid | BioGRID | 16169070 |
| RGL2 | HKE1.5 | KE1.5 | RAB2L | ral guanine nucleotide dissociation stimulator-like 2 | Two-hybrid | BioGRID | 16169070 |
| RUVBL1 | ECP54 | INO80H | NMP238 | PONTIN | Pontin52 | RVB1 | TIH1 | TIP49 | TIP49A | RuvB-like 1 (E. coli) | HTATIP (Tip60) interacts with RUVBL1 (pontin). | BIND | 15829968 |
| SAT1 | DC21 | KFSD | SAT | SSAT | SSAT-1 | spermidine/spermine N1-acetyltransferase 1 | Two-hybrid | BioGRID | 16169070 |
| SNAPIN | SNAPAP | SNAP-associated protein | Two-hybrid | BioGRID | 16169070 |
| SNRPD2 | SMD2 | SNRPD1 | small nuclear ribonucleoprotein D2 polypeptide 16.5kDa | Two-hybrid | BioGRID | 16169070 |
| STAT3 | APRF | FLJ20882 | HIES | MGC16063 | signal transducer and activator of transcription 3 (acute-phase response factor) | - | HPRD,BioGRID | 12551922 |
| SYN1 | SYN1a | SYN1b | SYNI | synapsin I | Two-hybrid | BioGRID | 16169070 |
| TRIB3 | C20orf97 | NIPK | SINK | SKIP3 | TRB3 | tribbles homolog 3 (Drosophila) | Two-hybrid | BioGRID | 16169070 |
| ZEB1 | AREB6 | BZP | DELTA-EF1 | MGC133261 | NIL-2-A | NIL-2A | NIL2A | TCF8 | ZEB | ZFHEP | ZFHX1A | zinc finger E-box binding homeobox 1 | - | HPRD,BioGRID | 11275565 |
| ZNF24 | KOX17 | RSG-A | ZNF191 | ZSCAN3 | Zfp191 | zinc finger protein 24 | Two-hybrid | BioGRID | 16169070 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| ST ERK1 ERK2 MAPK PATHWAY | 32 | 25 | All SZGR 2.0 genes in this pathway |
| ST MYOCYTE AD PATHWAY | 27 | 25 | All SZGR 2.0 genes in this pathway |
| PID P73PATHWAY | 79 | 59 | All SZGR 2.0 genes in this pathway |
| PID MYC ACTIV PATHWAY | 79 | 62 | All SZGR 2.0 genes in this pathway |
| PID ATM PATHWAY | 34 | 25 | All SZGR 2.0 genes in this pathway |
| PID MYC PATHWAY | 25 | 22 | All SZGR 2.0 genes in this pathway |
| PID AR TF PATHWAY | 53 | 38 | All SZGR 2.0 genes in this pathway |
| PID ATF2 PATHWAY | 59 | 43 | All SZGR 2.0 genes in this pathway |
| PID P53 REGULATION PATHWAY | 59 | 50 | All SZGR 2.0 genes in this pathway |
| WANG LMO4 TARGETS DN | 352 | 225 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA UP | 1821 | 933 | All SZGR 2.0 genes in this pathway |
| MARTIN INTERACT WITH HDAC | 44 | 31 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER SERUM RESPONSE AUGMENTED BY MYC | 108 | 74 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN UP | 479 | 299 | All SZGR 2.0 genes in this pathway |
| THEILGAARD NEUTROPHIL AT SKIN WOUND UP | 77 | 52 | All SZGR 2.0 genes in this pathway |
| DEBIASI APOPTOSIS BY REOVIRUS INFECTION UP | 314 | 201 | All SZGR 2.0 genes in this pathway |
| KAYO CALORIE RESTRICTION MUSCLE DN | 87 | 59 | All SZGR 2.0 genes in this pathway |
| WELCSH BRCA1 TARGETS UP | 198 | 132 | All SZGR 2.0 genes in this pathway |
| DOUGLAS BMI1 TARGETS UP | 566 | 371 | All SZGR 2.0 genes in this pathway |
| CHANDRAN METASTASIS DN | 306 | 191 | All SZGR 2.0 genes in this pathway |