Gene Page: ARPC1A
Summary ?
| GeneID | 10552 |
| Symbol | ARPC1A |
| Synonyms | Arc40|HEL-68|HEL-S-307|SOP2Hs|SOP2L |
| Description | actin related protein 2/3 complex subunit 1A |
| Reference | MIM:604220|HGNC:HGNC:703|Ensembl:ENSG00000241685|HPRD:07252|Vega:OTTHUMG00000154553 |
| Gene type | protein-coding |
| Map location | 7q22.1 |
| Pascal p-value | 0.79 |
| Sherlock p-value | 0.923 |
| Fetal beta | 0.378 |
| DMG | 1 (# studies) |
| Support | STRUCTURAL PLASTICITY G2Cdb.humanPSD G2Cdb.humanPSP |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg04828509 | 7 | 98923380 | ARPC1A | 3.18E-6 | -0.457 | 0.009 | DMG:Wockner_2014 |
Section II. Transcriptome annotation
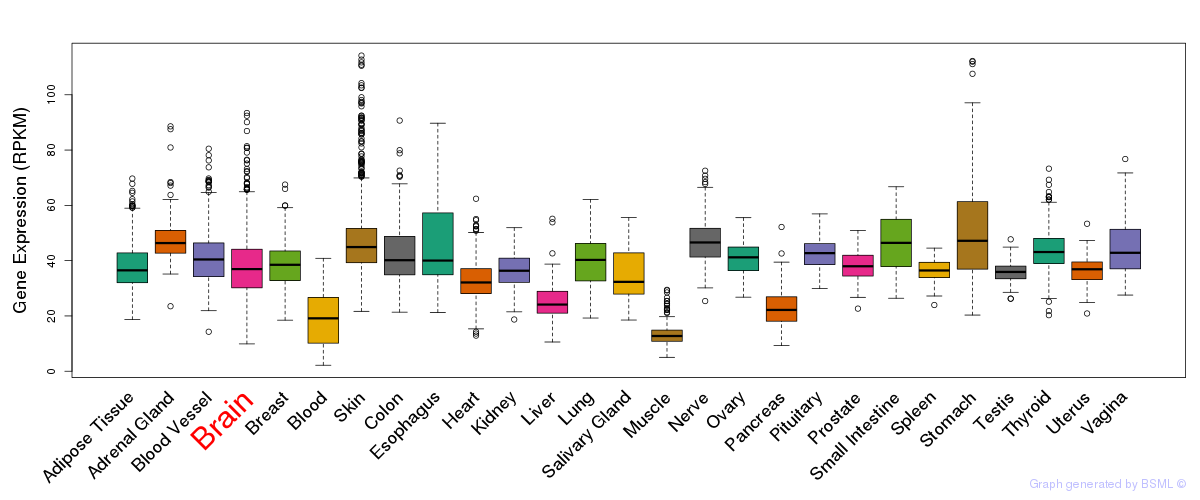
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| GPR26 | 0.67 | 0.62 |
| ARHGAP10 | 0.65 | 0.53 |
| SYP | 0.65 | 0.64 |
| LRRC53 | 0.63 | 0.47 |
| C1orf183 | 0.62 | 0.57 |
| PTPRR | 0.62 | 0.60 |
| VIT | 0.62 | 0.50 |
| STYK1 | 0.62 | 0.63 |
| ATF7IP2 | 0.62 | 0.56 |
| ARC | 0.62 | 0.50 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| RPL23A | -0.32 | -0.35 |
| SH2D2A | -0.31 | -0.33 |
| BCL7C | -0.28 | -0.32 |
| GPR125 | -0.28 | -0.25 |
| SIGIRR | -0.27 | -0.32 |
| TP53BP2 | -0.27 | -0.19 |
| AC132872.1 | -0.27 | -0.29 |
| CDC42EP4 | -0.27 | -0.21 |
| C21orf57 | -0.27 | -0.26 |
| TRAF4 | -0.27 | -0.29 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG FC GAMMA R MEDIATED PHAGOCYTOSIS | 97 | 71 | All SZGR 2.0 genes in this pathway |
| KEGG REGULATION OF ACTIN CYTOSKELETON | 216 | 144 | All SZGR 2.0 genes in this pathway |
| KEGG PATHOGENIC ESCHERICHIA COLI INFECTION | 59 | 36 | All SZGR 2.0 genes in this pathway |
| BIOCARTA SALMONELLA PATHWAY | 13 | 10 | All SZGR 2.0 genes in this pathway |
| BIOCARTA MPR PATHWAY | 34 | 28 | All SZGR 2.0 genes in this pathway |
| BIOCARTA RHO PATHWAY | 32 | 23 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CDC42RAC PATHWAY | 16 | 13 | All SZGR 2.0 genes in this pathway |
| BIOCARTA ACTINY PATHWAY | 20 | 14 | All SZGR 2.0 genes in this pathway |
| LIU SOX4 TARGETS DN | 309 | 191 | All SZGR 2.0 genes in this pathway |
| IGARASHI ATF4 TARGETS DN | 90 | 65 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| HAHTOLA MYCOSIS FUNGOIDES SKIN UP | 177 | 113 | All SZGR 2.0 genes in this pathway |
| HAHTOLA MYCOSIS FUNGOIDES CD4 UP | 64 | 46 | All SZGR 2.0 genes in this pathway |
| PROVENZANI METASTASIS DN | 136 | 94 | All SZGR 2.0 genes in this pathway |
| OLSSON E2F3 TARGETS UP | 28 | 14 | All SZGR 2.0 genes in this pathway |
| DAIRKEE TERT TARGETS UP | 380 | 213 | All SZGR 2.0 genes in this pathway |
| KOYAMA SEMA3B TARGETS DN | 411 | 249 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY BREAST CANCER 7Q21 Q22 AMPLICON | 76 | 33 | All SZGR 2.0 genes in this pathway |
| PARK HSC MARKERS | 44 | 31 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| WELCSH BRCA1 TARGETS UP | 198 | 132 | All SZGR 2.0 genes in this pathway |
| JIANG HYPOXIA NORMAL | 311 | 205 | All SZGR 2.0 genes in this pathway |
| AGUIRRE PANCREATIC CANCER COPY NUMBER UP | 298 | 174 | All SZGR 2.0 genes in this pathway |
| ZHAN LATE DIFFERENTIATION GENES UP | 33 | 24 | All SZGR 2.0 genes in this pathway |
| BOYAULT LIVER CANCER SUBCLASS G23 UP | 52 | 35 | All SZGR 2.0 genes in this pathway |
| CHIANG LIVER CANCER SUBCLASS UNANNOTATED DN | 193 | 112 | All SZGR 2.0 genes in this pathway |
| HAHTOLA CTCL CUTANEOUS | 26 | 19 | All SZGR 2.0 genes in this pathway |
| KYNG WERNER SYNDROM AND NORMAL AGING DN | 225 | 124 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 12 | 79 | 54 | All SZGR 2.0 genes in this pathway |
| SASSON RESPONSE TO GONADOTROPHINS UP | 91 | 59 | All SZGR 2.0 genes in this pathway |
| SASSON RESPONSE TO FORSKOLIN UP | 90 | 58 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS CALB1 CORR UP | 548 | 370 | All SZGR 2.0 genes in this pathway |
| HUANG GATA2 TARGETS UP | 149 | 96 | All SZGR 2.0 genes in this pathway |
| FORTSCHEGGER PHF8 TARGETS DN | 784 | 464 | All SZGR 2.0 genes in this pathway |
| LIM MAMMARY STEM CELL DN | 428 | 246 | All SZGR 2.0 genes in this pathway |