Gene Page: KHDRBS3
Summary ?
| GeneID | 10656 |
| Symbol | KHDRBS3 |
| Synonyms | Etle|SALP|SLM-2|SLM2|T-STAR|TSTAR|etoile |
| Description | KH domain containing, RNA binding, signal transduction associated 3 |
| Reference | MIM:610421|HGNC:HGNC:18117|Ensembl:ENSG00000131773|HPRD:10007|Vega:OTTHUMG00000164164 |
| Gene type | protein-coding |
| Map location | 8q24.2 |
| Pascal p-value | 0.207 |
| Sherlock p-value | 0.488 |
| DMG | 1 (# studies) |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 2 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg18587984 | 8 | 136473366 | KHDRBS3 | 4.74E-5 | 0.563 | 0.021 | DMG:Wockner_2014 |
| cg21933359 | 8 | 136531649 | KHDRBS3 | 3.017E-4 | 0.524 | 0.04 | DMG:Wockner_2014 |
Section II. Transcriptome annotation
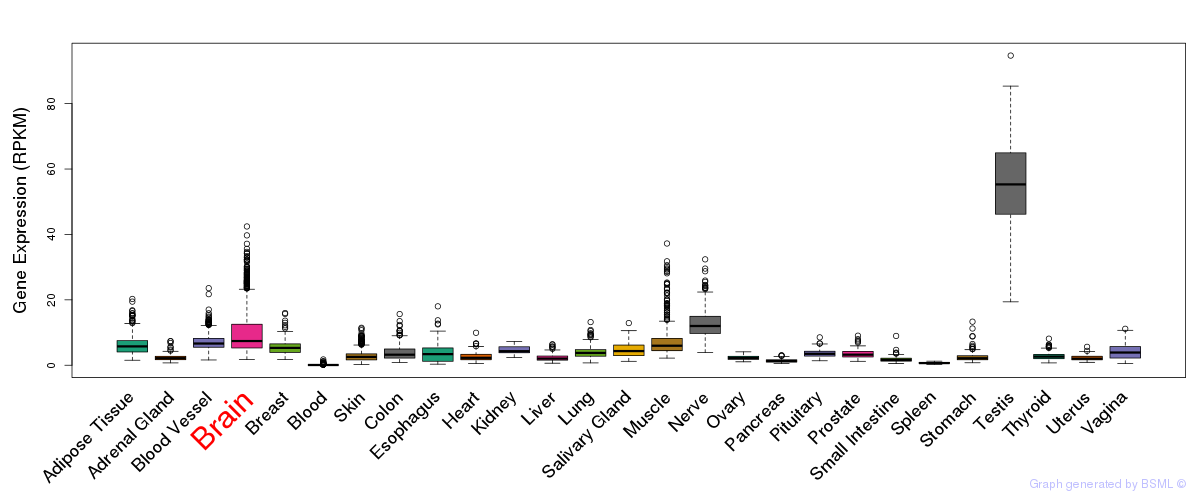
General gene expression (GTEx)
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| CBX6 | 0.83 | 0.82 |
| C2orf55 | 0.82 | 0.78 |
| IQSEC2 | 0.82 | 0.85 |
| SYNPO | 0.81 | 0.80 |
| MINK1 | 0.81 | 0.82 |
| KIAA1217 | 0.80 | 0.84 |
| KLHL34 | 0.80 | 0.76 |
| SPRYD3 | 0.79 | 0.79 |
| PACSIN1 | 0.79 | 0.78 |
| CAMTA2 | 0.79 | 0.81 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| BCL7C | -0.49 | -0.58 |
| HEBP2 | -0.47 | -0.60 |
| RPL23A | -0.44 | -0.49 |
| HYAL2 | -0.43 | -0.45 |
| RPS13P2 | -0.43 | -0.44 |
| RPL32 | -0.43 | -0.44 |
| RPS18 | -0.42 | -0.44 |
| C21orf57 | -0.42 | -0.49 |
| RPS23 | -0.42 | -0.44 |
| C9orf46 | -0.42 | -0.44 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| BAHD1 | KIAA0945 | bromo adjacent homology domain containing 1 | Two-hybrid | BioGRID | 16189514 |
| CCDC33 | FLJ23168 | FLJ32855 | MGC34145 | coiled-coil domain containing 33 | Two-hybrid | BioGRID | 16189514 |
| HNRNPK | CSBP | FLJ41122 | HNRPK | TUNP | heterogeneous nuclear ribonucleoprotein K | Two-hybrid | BioGRID | 16189514 |
| KHDRBS1 | FLJ34027 | Sam68 | p62 | KH domain containing, RNA binding, signal transduction associated 1 | - | HPRD,BioGRID | 10749975 |
| KHDRBS2 | FLJ38664 | MGC26664 | SLM-1 | SLM1 | bA535F17.1 | KH domain containing, RNA binding, signal transduction associated 2 | Two-hybrid | BioGRID | 16189514 |
| KHDRBS3 | Etle | SALP | SLM-2 | SLM2 | T-STAR | TSTAR | etoile | KH domain containing, RNA binding, signal transduction associated 3 | Two-hybrid | BioGRID | 10749975 |
| LNX1 | LNX | MPDZ | PDZRN2 | ligand of numb-protein X 1 | Two-hybrid | BioGRID | 16189514 |
| NUDT18 | FLJ22494 | nudix (nucleoside diphosphate linked moiety X)-type motif 18 | Two-hybrid | BioGRID | 16189514 |
| PRMT1 | ANM1 | HCP1 | HRMT1L2 | IR1B4 | protein arginine methyltransferase 1 | - | HPRD,BioGRID | 12529443 |
| PSMF1 | PI31 | proteasome (prosome, macropain) inhibitor subunit 1 (PI31) | Two-hybrid | BioGRID | 16189514 |
| RBM7 | FLJ11153 | RNA binding motif protein 7 | Two-hybrid | BioGRID | 16189514 |
| RBMX | HNRPG | RBMXP1 | RBMXRT | RNMX | hnRNP-G | RNA binding motif protein, X-linked | Two-hybrid | BioGRID | 16189514 |
| RBMY1A1 | MGC181956 | RBM1 | RBM2 | RBMY | YRRM1 | YRRM2 | RNA binding motif protein, Y-linked, family 1, member A1 | Two-hybrid | BioGRID | 10749975 |
| SFRS9 | SRp30c | splicing factor, arginine/serine-rich 9 | - | HPRD | 11118435 |
| SGSM2 | KIAA0397 | RUTBC1 | small G protein signaling modulator 2 | Two-hybrid | BioGRID | 16169070 |
| SIAH1 | FLJ08065 | HUMSIAH | Siah-1 | Siah-1a | hSIAH1 | seven in absentia homolog 1 (Drosophila) | - | HPRD,BioGRID | 15163637 |
| YTHDC1 | KIAA1966 | YT521 | YT521-B | YTH domain containing 1 | - | HPRD | 11118435 |
| ZNF408 | FLJ12827 | zinc finger protein 408 | Two-hybrid | BioGRID | 16189514 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| HOEBEKE LYMPHOID STEM CELL DN | 86 | 59 | All SZGR 2.0 genes in this pathway |
| AKL HTLV1 INFECTION DN | 68 | 41 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC3 TARGETS UP | 501 | 327 | All SZGR 2.0 genes in this pathway |
| JAATINEN HEMATOPOIETIC STEM CELL UP | 316 | 190 | All SZGR 2.0 genes in this pathway |
| PEREZ TP63 TARGETS | 355 | 243 | All SZGR 2.0 genes in this pathway |
| FURUKAWA DUSP6 TARGETS PCI35 DN | 74 | 40 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 6HR DN | 514 | 330 | All SZGR 2.0 genes in this pathway |
| KOYAMA SEMA3B TARGETS DN | 411 | 249 | All SZGR 2.0 genes in this pathway |
| GEORGES TARGETS OF MIR192 AND MIR215 | 893 | 528 | All SZGR 2.0 genes in this pathway |
| BECKER TAMOXIFEN RESISTANCE DN | 52 | 37 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 20HR UP | 240 | 152 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 16HR UP | 225 | 139 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS STEM CELL AND PROGENITOR | 681 | 420 | All SZGR 2.0 genes in this pathway |
| JIANG HYPOXIA NORMAL | 311 | 205 | All SZGR 2.0 genes in this pathway |
| SATO SILENCED BY METHYLATION IN PANCREATIC CANCER 1 | 419 | 273 | All SZGR 2.0 genes in this pathway |
| BILD E2F3 ONCOGENIC SIGNATURE | 246 | 153 | All SZGR 2.0 genes in this pathway |
| ACEVEDO NORMAL TISSUE ADJACENT TO LIVER TUMOR DN | 354 | 216 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER RELAPSE IN BONE DN | 315 | 197 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER LUMINAL B DN | 564 | 326 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL UP | 648 | 398 | All SZGR 2.0 genes in this pathway |
| IZADPANAH STEM CELL ADIPOSE VS BONE UP | 126 | 92 | All SZGR 2.0 genes in this pathway |
| LIN TUMOR ESCAPE FROM IMMUNE ATTACK | 18 | 12 | All SZGR 2.0 genes in this pathway |
| TAYLOR METHYLATED IN ACUTE LYMPHOBLASTIC LEUKEMIA | 77 | 52 | All SZGR 2.0 genes in this pathway |
| HAN SATB1 TARGETS UP | 395 | 249 | All SZGR 2.0 genes in this pathway |
| COLINA TARGETS OF 4EBP1 AND 4EBP2 | 356 | 214 | All SZGR 2.0 genes in this pathway |
| SETLUR PROSTATE CANCER TMPRSS2 ERG FUSION UP | 67 | 48 | All SZGR 2.0 genes in this pathway |
| WONG ADULT TISSUE STEM MODULE | 721 | 492 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS CALB1 CORR UP | 548 | 370 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 1 DN | 63 | 39 | All SZGR 2.0 genes in this pathway |
| TORCHIA TARGETS OF EWSR1 FLI1 FUSION DN | 321 | 200 | All SZGR 2.0 genes in this pathway |
| PLASARI TGFB1 TARGETS 10HR UP | 199 | 143 | All SZGR 2.0 genes in this pathway |
| ACEVEDO FGFR1 TARGETS IN PROSTATE CANCER MODEL DN | 308 | 187 | All SZGR 2.0 genes in this pathway |