Gene Page: CAPN9
Summary ?
| GeneID | 10753 |
| Symbol | CAPN9 |
| Synonyms | GC36|nCL-4 |
| Description | calpain 9 |
| Reference | MIM:606401|HGNC:HGNC:1486|Ensembl:ENSG00000135773|HPRD:05909|Vega:OTTHUMG00000037779 |
| Gene type | protein-coding |
| Map location | 1q42.11-q42.3 |
| TADA p-value | 0.013 |
| Fetal beta | -0.136 |
| eGene | Caudate basal ganglia Cerebellum Cortex Frontal Cortex BA9 Putamen basal ganglia Meta |
| Support | CompositeSet |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DNM:Fromer_2014 | Whole Exome Sequencing analysis | This study reported a WES study of 623 schizophrenia trios, reporting DNMs using genomic DNA. |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| CAPN9 | chr1 | 230903336 | A | G | NM_006615 NM_016452 | p.196M>V p.196M>V | missense missense | Schizophrenia | DNM:Fromer_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs11579420 | 1 | 230896238 | CAPN9 | ENSG00000135773.8 | 2.40106E-7 | 0.01 | 13108 | gtex_brain_putamen_basal |
Section II. Transcriptome annotation
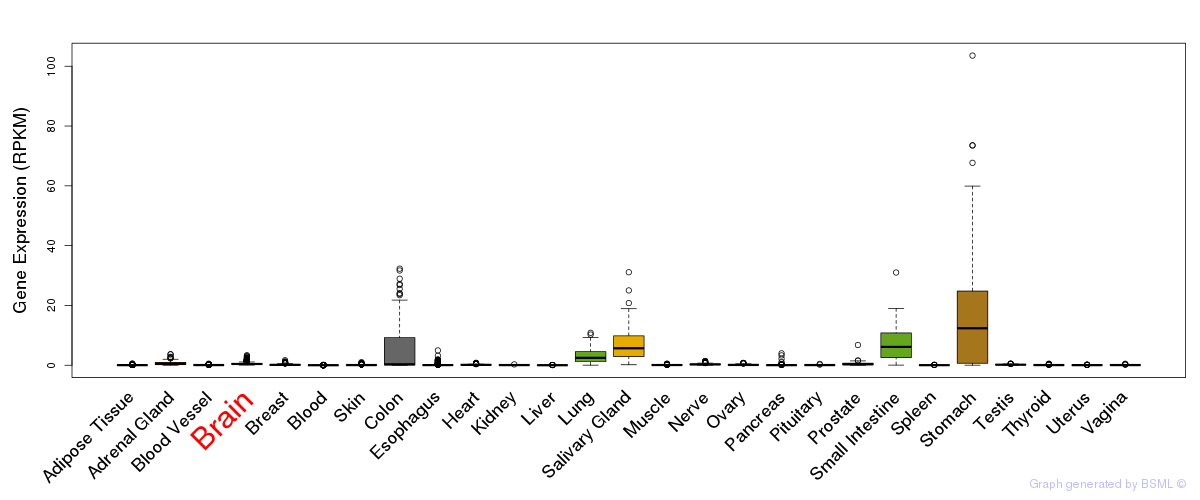
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| CHRNA1 | 0.56 | 0.28 |
| METTL7B | 0.48 | 0.29 |
| KCNJ13 | 0.47 | 0.27 |
| CP | 0.47 | 0.31 |
| CLIC6 | 0.46 | 0.31 |
| F5 | 0.46 | 0.34 |
| AQP1 | 0.46 | 0.38 |
| TMEM27 | 0.46 | 0.18 |
| PLBD1 | 0.45 | 0.29 |
| SLC5A5 | 0.44 | 0.31 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SH2D2A | -0.26 | -0.41 |
| SEMA4C | -0.26 | -0.34 |
| ZBTB12 | -0.25 | -0.30 |
| KIAA1949 | -0.25 | -0.28 |
| SH2B2 | -0.24 | -0.36 |
| SH3BP2 | -0.24 | -0.36 |
| FAM129B | -0.24 | -0.36 |
| SLC16A13 | -0.24 | -0.33 |
| FBXO46 | -0.24 | -0.31 |
| C21orf30 | -0.24 | -0.29 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| SENGUPTA NASOPHARYNGEAL CARCINOMA DN | 349 | 157 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS BASAL UP | 380 | 215 | All SZGR 2.0 genes in this pathway |
| VECCHI GASTRIC CANCER ADVANCED VS EARLY DN | 138 | 70 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA UP | 1821 | 933 | All SZGR 2.0 genes in this pathway |
| GOZGIT ESR1 TARGETS DN | 781 | 465 | All SZGR 2.0 genes in this pathway |
| FARMER BREAST CANCER BASAL VS LULMINAL | 330 | 215 | All SZGR 2.0 genes in this pathway |
| JAZAG TGFB1 SIGNALING UP | 108 | 69 | All SZGR 2.0 genes in this pathway |
| ABBUD LIF SIGNALING 1 DN | 26 | 17 | All SZGR 2.0 genes in this pathway |
| SANSOM APC TARGETS DN | 366 | 238 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 5 | 482 | 296 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER RELAPSE IN BONE UP | 97 | 61 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL DN | 701 | 446 | All SZGR 2.0 genes in this pathway |
| MARTENS TRETINOIN RESPONSE UP | 857 | 456 | All SZGR 2.0 genes in this pathway |
| FEVR CTNNB1 TARGETS UP | 682 | 433 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG RXRA BOUND 8D | 882 | 506 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 2ND EGF PULSE ONLY | 1725 | 838 | All SZGR 2.0 genes in this pathway |