Gene Page: SLC17A3
Summary ?
| GeneID | 10786 |
| Symbol | SLC17A3 |
| Synonyms | GOUT4|NPT4|UAQTL4 |
| Description | solute carrier family 17 member 3 |
| Reference | MIM:611034|HGNC:HGNC:10931|Ensembl:ENSG00000124564|HPRD:10233|Vega:OTTHUMG00000014412 |
| Gene type | protein-coding |
| Map location | 6p21.3 |
| Pascal p-value | 1E-12 |
| Fetal beta | 0.026 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| GSMA_I | Genome scan meta-analysis | Psr: 0.033 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
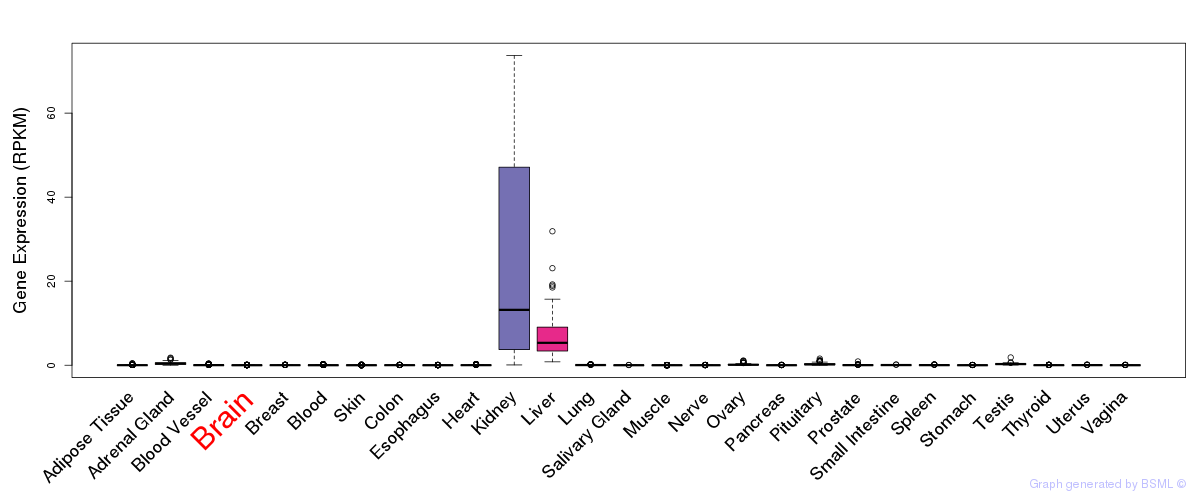
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| YWHAE | 0.93 | 0.92 |
| PTGES3 | 0.92 | 0.89 |
| EIF2S1 | 0.91 | 0.91 |
| DNAJB6 | 0.91 | 0.91 |
| DNAJC8 | 0.91 | 0.90 |
| TAF7 | 0.91 | 0.90 |
| GLOD4 | 0.90 | 0.88 |
| YEATS4 | 0.90 | 0.90 |
| DDX50 | 0.90 | 0.88 |
| C16orf94 | 0.90 | 0.87 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MT-CO2 | -0.71 | -0.70 |
| AF347015.2 | -0.70 | -0.71 |
| MT-CYB | -0.69 | -0.70 |
| AF347015.33 | -0.69 | -0.69 |
| AF347015.31 | -0.68 | -0.70 |
| AF347015.8 | -0.68 | -0.68 |
| AF347015.15 | -0.67 | -0.70 |
| AF347015.26 | -0.67 | -0.69 |
| AF347015.9 | -0.67 | -0.72 |
| AF347015.27 | -0.65 | -0.66 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005436 | sodium:phosphate symporter activity | TAS | 9149941 | |
| GO:0005215 | transporter activity | IEA | - | |
| GO:0015293 | symporter activity | IEA | - | |
| GO:0031402 | sodium ion binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006814 | sodium ion transport | TAS | 9149941 | |
| GO:0006811 | ion transport | IEA | - | |
| GO:0006810 | transport | IEA | - | |
| GO:0006796 | phosphate metabolic process | TAS | 9149941 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005789 | endoplasmic reticulum membrane | IEA | - | |
| GO:0005624 | membrane fraction | TAS | 9149941 | |
| GO:0005783 | endoplasmic reticulum | IEA | - | |
| GO:0016020 | membrane | IEA | - | |
| GO:0016021 | integral to membrane | IEA | - | |
| GO:0005887 | integral to plasma membrane | TAS | 9149941 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| LEE LIVER CANCER CIPROFIBRATE DN | 66 | 43 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT REJECTED VS OK DN | 546 | 351 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT OK VS DONOR UP | 555 | 346 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER UP | 973 | 570 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER TUMOR VS NORMAL ADJACENT TISSUE UP | 863 | 514 | All SZGR 2.0 genes in this pathway |
| ACEVEDO METHYLATED IN LIVER CANCER DN | 940 | 425 | All SZGR 2.0 genes in this pathway |
| OHGUCHI LIVER HNF4A TARGETS DN | 149 | 85 | All SZGR 2.0 genes in this pathway |
| SERVITJA LIVER HNF1A TARGETS DN | 157 | 105 | All SZGR 2.0 genes in this pathway |