Gene Page: PDE10A
Summary ?
| GeneID | 10846 |
| Symbol | PDE10A |
| Synonyms | HSPDE10A |
| Description | phosphodiesterase 10A |
| Reference | MIM:610652|HGNC:HGNC:8772|Ensembl:ENSG00000112541|HPRD:17828|Vega:OTTHUMG00000015986 |
| Gene type | protein-coding |
| Map location | 6q26 |
| Pascal p-value | 0.178 |
| Fetal beta | 1.888 |
| Support | G2Cdb.humanPSD G2Cdb.humanPSP |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
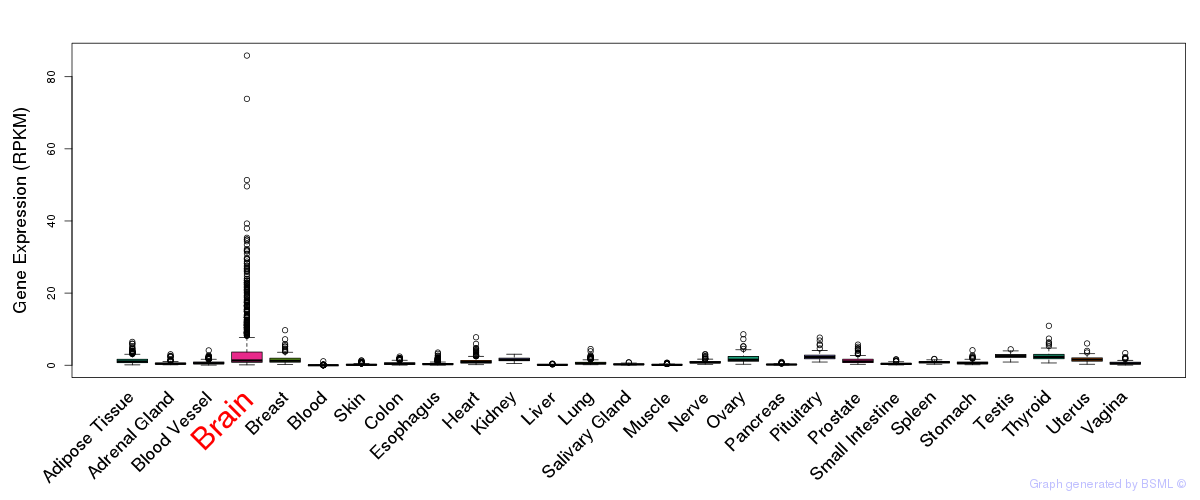
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| TACC3 | 0.99 | 0.72 |
| AURKB | 0.99 | 0.79 |
| FAM64A | 0.99 | 0.78 |
| KIF20A | 0.99 | 0.79 |
| PLK1 | 0.98 | 0.78 |
| NCAPH | 0.98 | 0.79 |
| UBE2C | 0.98 | 0.78 |
| HJURP | 0.98 | 0.79 |
| GTSE1 | 0.98 | 0.83 |
| NUSAP1 | 0.98 | 0.81 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| C5orf53 | -0.38 | -0.68 |
| FBXO2 | -0.38 | -0.63 |
| HLA-F | -0.38 | -0.70 |
| SLC9A3R2 | -0.37 | -0.41 |
| PTH1R | -0.37 | -0.61 |
| ASPHD1 | -0.37 | -0.56 |
| ALDOC | -0.36 | -0.60 |
| AIFM3 | -0.36 | -0.59 |
| AF347015.27 | -0.36 | -0.73 |
| TNFSF12 | -0.36 | -0.58 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0004114 | 3',5'-cyclic-nucleotide phosphodiesterase activity | IEA | - | |
| GO:0004114 | 3',5'-cyclic-nucleotide phosphodiesterase activity | TAS | 10373451 | |
| GO:0016787 | hydrolase activity | IEA | - | |
| GO:0030553 | cGMP binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007165 | signal transduction | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005737 | cytoplasm | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG PURINE METABOLISM | 159 | 96 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY GPCR | 920 | 449 | All SZGR 2.0 genes in this pathway |
| REACTOME GPCR DOWNSTREAM SIGNALING | 805 | 368 | All SZGR 2.0 genes in this pathway |
| REACTOME G ALPHA S SIGNALLING EVENTS | 121 | 82 | All SZGR 2.0 genes in this pathway |
| REACTOME CGMP EFFECTS | 19 | 13 | All SZGR 2.0 genes in this pathway |
| REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | 25 | 19 | All SZGR 2.0 genes in this pathway |
| REACTOME PLATELET HOMEOSTASIS | 78 | 49 | All SZGR 2.0 genes in this pathway |
| REACTOME HEMOSTASIS | 466 | 331 | All SZGR 2.0 genes in this pathway |
| ONKEN UVEAL MELANOMA UP | 783 | 507 | All SZGR 2.0 genes in this pathway |
| SCHUETZ BREAST CANCER DUCTAL INVASIVE UP | 351 | 230 | All SZGR 2.0 genes in this pathway |
| WATANABE COLON CANCER MSI VS MSS UP | 29 | 18 | All SZGR 2.0 genes in this pathway |
| GAZDA DIAMOND BLACKFAN ANEMIA PROGENITOR UP | 39 | 25 | All SZGR 2.0 genes in this pathway |
| CORRE MULTIPLE MYELOMA UP | 74 | 45 | All SZGR 2.0 genes in this pathway |
| PEPPER CHRONIC LYMPHOCYTIC LEUKEMIA DN | 21 | 8 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS UP | 214 | 155 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS 8HR UP | 164 | 122 | All SZGR 2.0 genes in this pathway |
| DELYS THYROID CANCER DN | 232 | 154 | All SZGR 2.0 genes in this pathway |
| GAUSSMANN MLL AF4 FUSION TARGETS E UP | 97 | 60 | All SZGR 2.0 genes in this pathway |
| PEREZ TP53 TARGETS | 1174 | 695 | All SZGR 2.0 genes in this pathway |
| SILIGAN BOUND BY EWS FLT1 FUSION | 47 | 34 | All SZGR 2.0 genes in this pathway |
| BUYTAERT PHOTODYNAMIC THERAPY STRESS UP | 811 | 508 | All SZGR 2.0 genes in this pathway |
| BENPORATH EED TARGETS | 1062 | 725 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES WITH H3K27ME3 | 1118 | 744 | All SZGR 2.0 genes in this pathway |
| KANG IMMORTALIZED BY TERT DN | 102 | 67 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 DN | 830 | 547 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS DN | 31 | 23 | All SZGR 2.0 genes in this pathway |
| YEGNASUBRAMANIAN PROSTATE CANCER | 128 | 60 | All SZGR 2.0 genes in this pathway |
| RIGGINS TAMOXIFEN RESISTANCE DN | 220 | 147 | All SZGR 2.0 genes in this pathway |
| NAKAMURA METASTASIS | 47 | 28 | All SZGR 2.0 genes in this pathway |
| NAKAMURA METASTASIS MODEL UP | 45 | 26 | All SZGR 2.0 genes in this pathway |
| HAN SATB1 TARGETS UP | 395 | 249 | All SZGR 2.0 genes in this pathway |
| HAN SATB1 TARGETS DN | 442 | 275 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K4ME3 AND H3K27ME3 | 1069 | 729 | All SZGR 2.0 genes in this pathway |
| NAKAMURA ADIPOGENESIS EARLY UP | 66 | 44 | All SZGR 2.0 genes in this pathway |
| NAKAMURA ADIPOGENESIS LATE UP | 104 | 67 | All SZGR 2.0 genes in this pathway |
| VERHAAK GLIOBLASTOMA PRONEURAL | 177 | 132 | All SZGR 2.0 genes in this pathway |
| PHONG TNF RESPONSE NOT VIA P38 | 337 | 236 | All SZGR 2.0 genes in this pathway |