Gene Page: TSPAN9
Summary ?
| GeneID | 10867 |
| Symbol | TSPAN9 |
| Synonyms | NET-5|NET5|PP1057 |
| Description | tetraspanin 9 |
| Reference | MIM:613137|HGNC:HGNC:21640|Ensembl:ENSG00000011105|HPRD:14823|Vega:OTTHUMG00000150333 |
| Gene type | protein-coding |
| Map location | 12p13.33-p13.32 |
| Pascal p-value | 0.64 |
| Fetal beta | -0.066 |
| DMG | 2 (# studies) |
| eGene | Cerebellar Hemisphere Putamen basal ganglia Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Nishioka_2013 | Genome-wide DNA methylation analysis | The authors investigated the methylation profiles of DNA in peripheral blood cells from 18 patients with first-episode schizophrenia (FESZ) and from 15 normal controls. | 3 |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 3 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg24281891 | 12 | 3194425 | TSPAN9 | 3.543E-4 | 0.256 | 0.042 | DMG:Wockner_2014 |
| cg07248242 | 12 | 3281520 | TSPAN9 | 4.576E-4 | -0.366 | 0.046 | DMG:Wockner_2014 |
| cg00081935 | 12 | 3186906 | TSPAN9 | -0.023 | 0.81 | DMG:Nishioka_2013 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs2502827 | chr1 | 176044216 | TSPAN9 | 10867 | 0.01 | trans | ||
| rs12405921 | chr1 | 206695730 | TSPAN9 | 10867 | 0.14 | trans | ||
| rs17572651 | chr1 | 218943612 | TSPAN9 | 10867 | 0.11 | trans | ||
| rs16829545 | chr2 | 151977407 | TSPAN9 | 10867 | 1.794E-10 | trans | ||
| rs3845734 | chr2 | 171125572 | TSPAN9 | 10867 | 1.584E-4 | trans | ||
| rs7584986 | chr2 | 184111432 | TSPAN9 | 10867 | 3.854E-8 | trans | ||
| rs17762315 | chr5 | 76807576 | TSPAN9 | 10867 | 0.04 | trans | ||
| rs1368303 | chr5 | 147672388 | TSPAN9 | 10867 | 0.04 | trans | ||
| rs1929769 | chr6 | 40765977 | TSPAN9 | 10867 | 0.18 | trans | ||
| rs2393316 | chr10 | 59333070 | TSPAN9 | 10867 | 0.16 | trans | ||
| rs17104720 | chr14 | 77127308 | TSPAN9 | 10867 | 0.05 | trans | ||
| rs16955618 | chr15 | 29937543 | TSPAN9 | 10867 | 3.11E-23 | trans | ||
| rs1041786 | chr21 | 22617710 | TSPAN9 | 10867 | 0 | trans | ||
| rs11062561 | 12 | 3305091 | TSPAN9 | ENSG00000011105.7 | 2.107E-6 | 0 | 118570 | gtex_brain_putamen_basal |
| rs11062562 | 12 | 3305394 | TSPAN9 | ENSG00000011105.7 | 2.098E-6 | 0 | 118873 | gtex_brain_putamen_basal |
| rs12372425 | 12 | 3305905 | TSPAN9 | ENSG00000011105.7 | 2.098E-6 | 0 | 119384 | gtex_brain_putamen_basal |
| rs73047986 | 12 | 3311182 | TSPAN9 | ENSG00000011105.7 | 2.098E-6 | 0 | 124661 | gtex_brain_putamen_basal |
| rs11062564 | 12 | 3313540 | TSPAN9 | ENSG00000011105.7 | 9.572E-8 | 0 | 127019 | gtex_brain_putamen_basal |
| rs7979334 | 12 | 3316270 | TSPAN9 | ENSG00000011105.7 | 9.579E-8 | 0 | 129749 | gtex_brain_putamen_basal |
| rs2335666 | 12 | 3318258 | TSPAN9 | ENSG00000011105.7 | 1.06E-6 | 0 | 131737 | gtex_brain_putamen_basal |
Section II. Transcriptome annotation
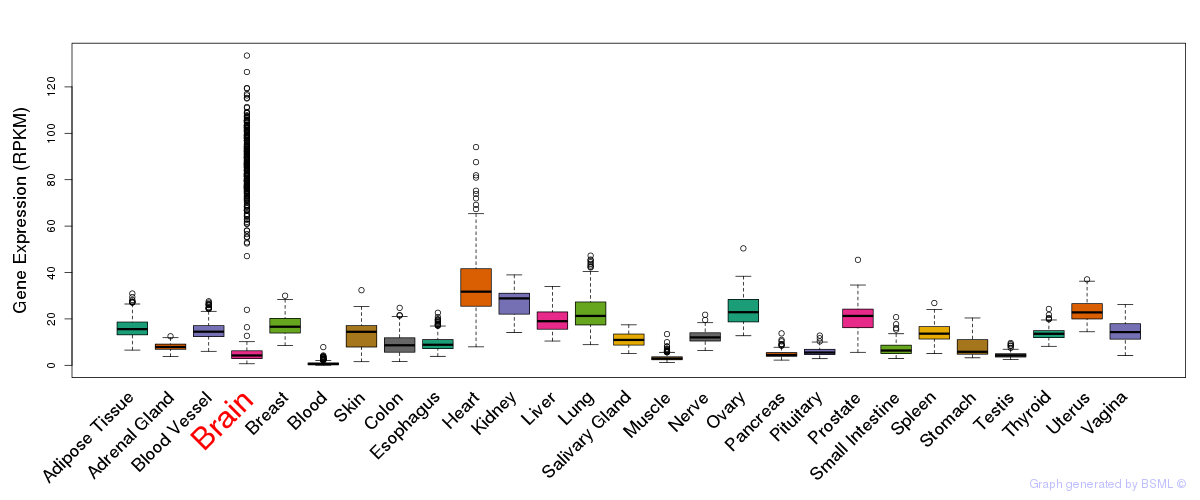
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| LILRA6 | 0.73 | 0.63 |
| FPR1 | 0.46 | 0.34 |
| CD14 | 0.46 | 0.28 |
| VSIG4 | 0.46 | 0.36 |
| CD68 | 0.45 | 0.34 |
| SLC37A2 | 0.45 | 0.31 |
| FCGR2A | 0.44 | 0.37 |
| SERPINA1 | 0.44 | 0.24 |
| SIGLEC14 | 0.43 | 0.37 |
| HMOX1 | 0.42 | 0.28 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| C5orf53 | -0.26 | -0.32 |
| C10orf4 | -0.25 | -0.30 |
| UQCRB | -0.24 | -0.32 |
| NDUFB3 | -0.22 | -0.30 |
| AC008670.2 | -0.22 | -0.38 |
| MAP3K15 | -0.22 | -0.26 |
| BXDC1 | -0.22 | -0.27 |
| SERGEF | -0.22 | -0.24 |
| AC104794.1 | -0.21 | -0.27 |
| SERPINI1 | -0.21 | -0.22 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KORKOLA SEMINOMA UP | 44 | 27 | All SZGR 2.0 genes in this pathway |
| DAVICIONI MOLECULAR ARMS VS ERMS DN | 182 | 111 | All SZGR 2.0 genes in this pathway |
| GAL LEUKEMIC STEM CELL UP | 133 | 78 | All SZGR 2.0 genes in this pathway |
| CONCANNON APOPTOSIS BY EPOXOMICIN UP | 239 | 157 | All SZGR 2.0 genes in this pathway |
| TERAMOTO OPN TARGETS CLUSTER 6 | 27 | 17 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS UP | 1037 | 673 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 24HR UP | 148 | 96 | All SZGR 2.0 genes in this pathway |
| LEE METASTASIS AND ALTERNATIVE SPLICING UP | 74 | 51 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER DN | 540 | 340 | All SZGR 2.0 genes in this pathway |
| KRISHNAN FURIN TARGETS DN | 13 | 8 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA HAPTOTAXIS DN | 668 | 419 | All SZGR 2.0 genes in this pathway |
| WIERENGA STAT5A TARGETS DN | 213 | 127 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS UP | 504 | 321 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP A | 898 | 516 | All SZGR 2.0 genes in this pathway |
| PASINI SUZ12 TARGETS DN | 315 | 215 | All SZGR 2.0 genes in this pathway |
| DELACROIX RARG BOUND MEF | 367 | 231 | All SZGR 2.0 genes in this pathway |
| DELACROIX RAR BOUND ES | 462 | 273 | All SZGR 2.0 genes in this pathway |
| RAO BOUND BY SALL4 ISOFORM B | 517 | 302 | All SZGR 2.0 genes in this pathway |