| KEGG ENDOCYTOSIS
| 183 | 132 | All SZGR 2.0 genes in this pathway |
| REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION
| 132 | 101 | All SZGR 2.0 genes in this pathway |
| REACTOME HEMOSTASIS
| 466 | 331 | All SZGR 2.0 genes in this pathway |
| ZHONG RESPONSE TO AZACITIDINE AND TSA UP
| 183 | 119 | All SZGR 2.0 genes in this pathway |
| DAVICIONI MOLECULAR ARMS VS ERMS DN
| 182 | 111 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE LIVE UP
| 485 | 293 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP
| 1382 | 904 | All SZGR 2.0 genes in this pathway |
| LIU TARGETS OF VMYB VS CMYB DN
| 43 | 30 | All SZGR 2.0 genes in this pathway |
| OSWALD HEMATOPOIETIC STEM CELL IN COLLAGEN GEL UP
| 233 | 161 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN NPM1 MUTATED SIGNATURE 1 DN
| 126 | 86 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN NPM1 MUTATED SIGNATURE 2 DN
| 77 | 46 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN NPM1 SIGNATURE 3 DN
| 162 | 116 | All SZGR 2.0 genes in this pathway |
| UDAYAKUMAR MED1 TARGETS DN
| 240 | 171 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE FIMA UP
| 544 | 308 | All SZGR 2.0 genes in this pathway |
| SCIBETTA KDM5B TARGETS UP
| 19 | 12 | All SZGR 2.0 genes in this pathway |
| BIDUS METASTASIS DN
| 161 | 93 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY SERUM DEPRIVATION DN
| 234 | 147 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN UP
| 1142 | 669 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 4 5WK DN
| 196 | 131 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 6 7WK UP
| 197 | 135 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL TGFB1 TARGETS UP
| 169 | 127 | All SZGR 2.0 genes in this pathway |
| XU HGF SIGNALING NOT VIA AKT1 6HR
| 26 | 15 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS UP
| 1037 | 673 | All SZGR 2.0 genes in this pathway |
| RICKMAN TUMOR DIFFERENTIATED WELL VS POORLY DN
| 382 | 224 | All SZGR 2.0 genes in this pathway |
| AMIT EGF RESPONSE 240 HELA
| 60 | 43 | All SZGR 2.0 genes in this pathway |
| AMIT EGF RESPONSE 480 HELA
| 164 | 118 | All SZGR 2.0 genes in this pathway |
| ZHANG RESPONSE TO IKK INHIBITOR AND TNF UP
| 223 | 140 | All SZGR 2.0 genes in this pathway |
| MORI SMALL PRE BII LYMPHOCYTE DN
| 76 | 52 | All SZGR 2.0 genes in this pathway |
| ROZANOV MMP14 TARGETS DN
| 35 | 22 | All SZGR 2.0 genes in this pathway |
| PENG GLUCOSE DEPRIVATION DN
| 169 | 112 | All SZGR 2.0 genes in this pathway |
| GALINDO IMMUNE RESPONSE TO ENTEROTOXIN
| 85 | 67 | All SZGR 2.0 genes in this pathway |
| HESS TARGETS OF HOXA9 AND MEIS1 DN
| 77 | 48 | All SZGR 2.0 genes in this pathway |
| AFFAR YY1 TARGETS UP
| 214 | 133 | All SZGR 2.0 genes in this pathway |
| BASSO CD40 SIGNALING UP
| 101 | 76 | All SZGR 2.0 genes in this pathway |
| REN ALVEOLAR RHABDOMYOSARCOMA DN
| 408 | 274 | All SZGR 2.0 genes in this pathway |
| HEDENFALK BREAST CANCER BRACX DN
| 20 | 14 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP
| 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS 6
| 189 | 112 | All SZGR 2.0 genes in this pathway |
| BILD HRAS ONCOGENIC SIGNATURE
| 261 | 166 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR DN
| 911 | 527 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR UP
| 783 | 442 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 5
| 482 | 296 | All SZGR 2.0 genes in this pathway |
| QI PLASMACYTOMA UP
| 259 | 185 | All SZGR 2.0 genes in this pathway |
| WANG TUMOR INVASIVENESS UP
| 374 | 247 | All SZGR 2.0 genes in this pathway |
| SWEET LUNG CANCER KRAS UP
| 491 | 316 | All SZGR 2.0 genes in this pathway |
| HOFFMANN SMALL PRE BII TO IMMATURE B LYMPHOCYTE UP
| 70 | 49 | All SZGR 2.0 genes in this pathway |
| FOURNIER ACINAR DEVELOPMENT LATE 2
| 277 | 172 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS CONFLUENT
| 567 | 365 | All SZGR 2.0 genes in this pathway |
| WANG RESPONSE TO GSK3 INHIBITOR SB216763 DN
| 374 | 217 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 3 UP
| 430 | 288 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS DN
| 882 | 538 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION DN
| 1080 | 713 | All SZGR 2.0 genes in this pathway |
| PEDERSEN METASTASIS BY ERBB2 ISOFORM 7
| 403 | 240 | All SZGR 2.0 genes in this pathway |
| KRIEG KDM3A TARGETS NOT HYPOXIA
| 208 | 107 | All SZGR 2.0 genes in this pathway |
| ALFANO MYC TARGETS
| 239 | 156 | All SZGR 2.0 genes in this pathway |
| FORTSCHEGGER PHF8 TARGETS DN
| 784 | 464 | All SZGR 2.0 genes in this pathway |
| PHONG TNF RESPONSE NOT VIA P38
| 337 | 236 | All SZGR 2.0 genes in this pathway |
| ZWANG CLASS 1 TRANSIENTLY INDUCED BY EGF
| 516 | 308 | All SZGR 2.0 genes in this pathway |
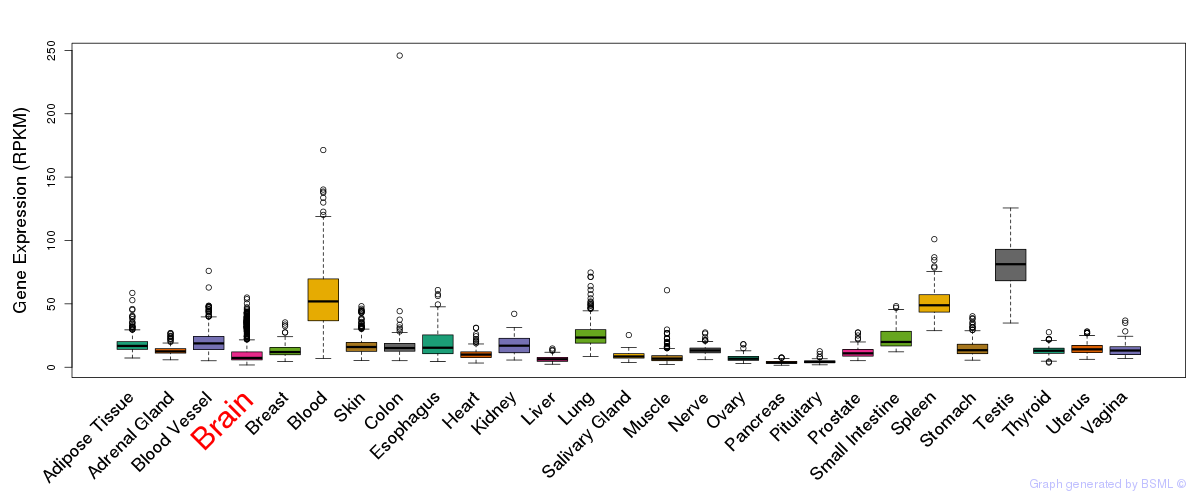
 Differentially methylated gene
Differentially methylated gene eQTL annotation
eQTL annotation