Gene Page: AFG3L2
Summary ?
| GeneID | 10939 |
| Symbol | AFG3L2 |
| Synonyms | SCA28|SPAX5 |
| Description | AFG3 like matrix AAA peptidase subunit 2 |
| Reference | MIM:604581|HGNC:HGNC:315|Ensembl:ENSG00000141385|HPRD:05205|Vega:OTTHUMG00000131695 |
| Gene type | protein-coding |
| Map location | 18p11 |
| Pascal p-value | 0.721 |
| Sherlock p-value | 0.68 |
| Fetal beta | -0.769 |
| DMG | 1 (# studies) |
| Support | Ascano FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg20012885 | 18 | 12377404 | AFG3L2 | 9.33E-5 | -0.28 | 0.027 | DMG:Wockner_2014 |
Section II. Transcriptome annotation
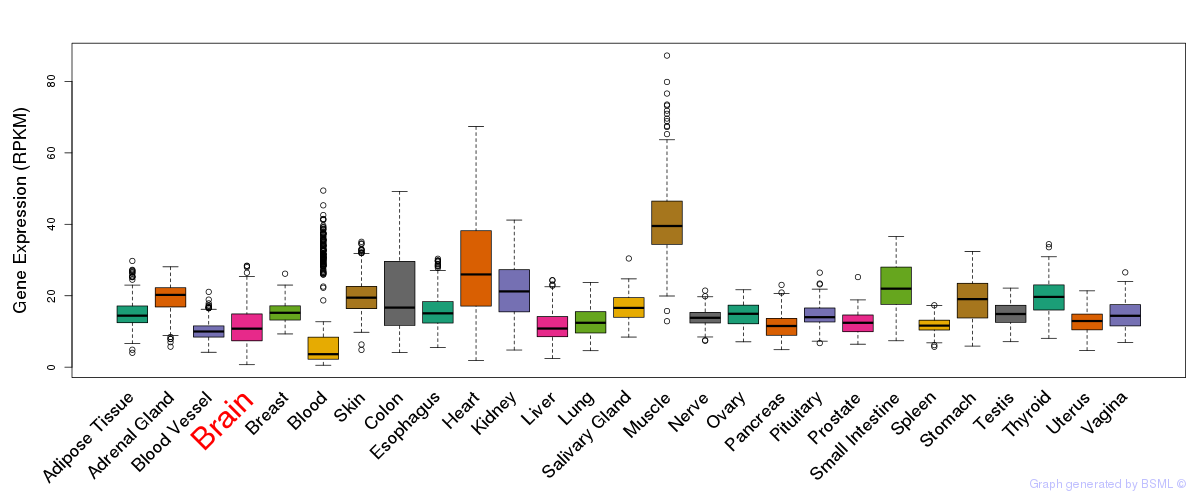
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| C3orf59 | 0.85 | 0.75 |
| PTPRO | 0.82 | 0.73 |
| FZD4 | 0.82 | 0.67 |
| CSRP2 | 0.80 | 0.57 |
| SDK1 | 0.79 | 0.70 |
| PDE4D | 0.79 | 0.72 |
| GLRA2 | 0.77 | 0.70 |
| STK32B | 0.77 | 0.71 |
| VLDLR | 0.77 | 0.68 |
| PANX1 | 0.77 | 0.74 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| C5orf53 | -0.55 | -0.66 |
| SLC16A11 | -0.53 | -0.57 |
| HEPN1 | -0.53 | -0.59 |
| LHPP | -0.52 | -0.54 |
| RGN | -0.52 | -0.57 |
| AC018755.7 | -0.52 | -0.59 |
| HLA-F | -0.51 | -0.58 |
| TSC22D4 | -0.51 | -0.59 |
| S100B | -0.51 | -0.64 |
| PACSIN3 | -0.51 | -0.53 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PUIFFE INVASION INHIBITED BY ASCITES DN | 145 | 91 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE KERATINOCYTE DN | 485 | 334 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER MYC TARGETS REPRESSED BY SERUM | 159 | 93 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| GROSS HYPOXIA VIA ELK3 AND HIF1A UP | 142 | 104 | All SZGR 2.0 genes in this pathway |
| BASSO B LYMPHOCYTE NETWORK | 143 | 96 | All SZGR 2.0 genes in this pathway |
| CROMER METASTASIS DN | 81 | 58 | All SZGR 2.0 genes in this pathway |
| KAAB HEART ATRIUM VS VENTRICLE DN | 261 | 183 | All SZGR 2.0 genes in this pathway |
| STEIN ESRRA TARGETS UP | 388 | 234 | All SZGR 2.0 genes in this pathway |
| TOYOTA TARGETS OF MIR34B AND MIR34C | 463 | 262 | All SZGR 2.0 genes in this pathway |
| MOOTHA HUMAN MITODB 6 2002 | 429 | 260 | All SZGR 2.0 genes in this pathway |
| MOOTHA MITOCHONDRIA | 447 | 277 | All SZGR 2.0 genes in this pathway |
| STEIN ESRRA TARGETS | 535 | 325 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS OLIGODENDROCYTE NUMBER CORR UP | 756 | 494 | All SZGR 2.0 genes in this pathway |