Gene Page: CKAP4
Summary ?
| GeneID | 10970 |
| Symbol | CKAP4 |
| Synonyms | CLIMP-63|ERGIC-63|p63 |
| Description | cytoskeleton-associated protein 4 |
| Reference | HGNC:HGNC:16991|Ensembl:ENSG00000136026|HPRD:16717|Vega:OTTHUMG00000170085 |
| Gene type | protein-coding |
| Map location | 12q23.3 |
| Pascal p-value | 0.096 |
| Sherlock p-value | 0.008 |
| Fetal beta | 0.491 |
| DMG | 1 (# studies) |
| Support | G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-CONSENSUS G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL G2Cdb.human_BAYES-COLLINS-MOUSE-PSD-CONSENSUS CompositeSet Ascano FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:vanEijk_2014 | Genome-wide DNA methylation analysis | This dataset includes 432 differentially methylated CpG sites corresponding to 391 unique transcripts between schizophrenia patients (n=260) and unaffected controls (n=250). | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg23555120 | 12 | 106533863 | CKAP4 | 0.007 | -4.301 | DMG:vanEijk_2014 |
Section II. Transcriptome annotation
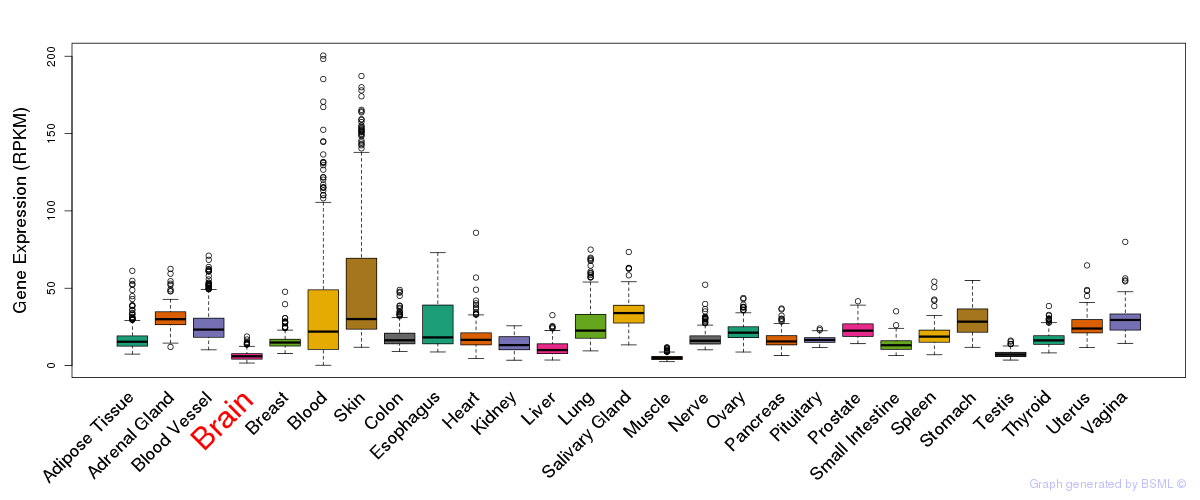
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SFRS8 | 0.90 | 0.90 |
| SNRNP70 | 0.88 | 0.85 |
| ZNF276 | 0.87 | 0.91 |
| GIGYF1 | 0.86 | 0.88 |
| LRDD | 0.86 | 0.88 |
| PABPN1 | 0.85 | 0.83 |
| ANKS3 | 0.85 | 0.86 |
| UCKL1 | 0.84 | 0.84 |
| TRABD | 0.84 | 0.86 |
| LRCH4 | 0.84 | 0.87 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.31 | -0.60 | -0.64 |
| B2M | -0.58 | -0.60 |
| GIMAP2 | -0.55 | -0.60 |
| AF347015.27 | -0.54 | -0.59 |
| C5orf53 | -0.53 | -0.53 |
| MT-CO2 | -0.53 | -0.59 |
| MT-CYB | -0.53 | -0.57 |
| EAF2 | -0.53 | -0.56 |
| ABCG2 | -0.52 | -0.55 |
| AF347015.21 | -0.51 | -0.65 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| HOLLMANN APOPTOSIS VIA CD40 UP | 201 | 125 | All SZGR 2.0 genes in this pathway |
| SENGUPTA NASOPHARYNGEAL CARCINOMA UP | 294 | 178 | All SZGR 2.0 genes in this pathway |
| CASORELLI ACUTE PROMYELOCYTIC LEUKEMIA DN | 663 | 425 | All SZGR 2.0 genes in this pathway |
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION HSC DN | 187 | 115 | All SZGR 2.0 genes in this pathway |
| TIEN INTESTINE PROBIOTICS 2HR DN | 88 | 53 | All SZGR 2.0 genes in this pathway |
| TIEN INTESTINE PROBIOTICS 6HR DN | 167 | 100 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA DN | 1375 | 806 | All SZGR 2.0 genes in this pathway |
| GOZGIT ESR1 TARGETS DN | 781 | 465 | All SZGR 2.0 genes in this pathway |
| TOMIDA METASTASIS UP | 26 | 13 | All SZGR 2.0 genes in this pathway |
| LI AMPLIFIED IN LUNG CANCER | 178 | 108 | All SZGR 2.0 genes in this pathway |
| MARTORIATI MDM4 TARGETS FETAL LIVER DN | 514 | 319 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN DN | 584 | 395 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS UP | 1037 | 673 | All SZGR 2.0 genes in this pathway |
| WEI MYCN TARGETS WITH E BOX | 795 | 478 | All SZGR 2.0 genes in this pathway |
| LIAO METASTASIS | 539 | 324 | All SZGR 2.0 genes in this pathway |
| PASQUALUCCI LYMPHOMA BY GC STAGE UP | 283 | 177 | All SZGR 2.0 genes in this pathway |
| GEORGES TARGETS OF MIR192 AND MIR215 | 893 | 528 | All SZGR 2.0 genes in this pathway |
| TARTE PLASMA CELL VS B LYMPHOCYTE UP | 78 | 51 | All SZGR 2.0 genes in this pathway |
| HALMOS CEBPA TARGETS UP | 52 | 34 | All SZGR 2.0 genes in this pathway |
| BENNETT SYSTEMIC LUPUS ERYTHEMATOSUS | 31 | 21 | All SZGR 2.0 genes in this pathway |
| JIANG VHL TARGETS | 138 | 91 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| RODWELL AGING KIDNEY NO BLOOD UP | 222 | 139 | All SZGR 2.0 genes in this pathway |
| KAYO CALORIE RESTRICTION MUSCLE UP | 95 | 64 | All SZGR 2.0 genes in this pathway |
| SESTO RESPONSE TO UV C7 | 68 | 44 | All SZGR 2.0 genes in this pathway |
| WELCSH BRCA1 TARGETS DN | 141 | 92 | All SZGR 2.0 genes in this pathway |
| AMUNDSON POOR SURVIVAL AFTER GAMMA RADIATION 8G | 95 | 62 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER TUMOR VS NORMAL ADJACENT TISSUE UP | 863 | 514 | All SZGR 2.0 genes in this pathway |
| CHEN HOXA5 TARGETS 9HR DN | 41 | 17 | All SZGR 2.0 genes in this pathway |
| PARK APL PATHOGENESIS DN | 50 | 35 | All SZGR 2.0 genes in this pathway |
| HOFMANN MYELODYSPLASTIC SYNDROM LOW RISK DN | 30 | 20 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA CHEMOTAXIS DN | 457 | 302 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA HAPTOTAXIS DN | 668 | 419 | All SZGR 2.0 genes in this pathway |
| BOYAULT LIVER CANCER SUBCLASS G123 UP | 45 | 21 | All SZGR 2.0 genes in this pathway |
| CHIANG LIVER CANCER SUBCLASS PROLIFERATION UP | 178 | 108 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA UP | 207 | 143 | All SZGR 2.0 genes in this pathway |
| JIANG HYPOXIA VIA VHL | 34 | 24 | All SZGR 2.0 genes in this pathway |
| WIERENGA STAT5A TARGETS UP | 217 | 131 | All SZGR 2.0 genes in this pathway |
| WIERENGA STAT5A TARGETS GROUP2 | 60 | 38 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |