Gene Page: CHD1
Summary ?
| GeneID | 1105 |
| Symbol | CHD1 |
| Synonyms | - |
| Description | chromodomain helicase DNA binding protein 1 |
| Reference | MIM:602118|HGNC:HGNC:1915|Ensembl:ENSG00000153922|HPRD:03668|Vega:OTTHUMG00000162744 |
| Gene type | protein-coding |
| Map location | 5q15-q21 |
| Pascal p-value | 0.005 |
| Sherlock p-value | 0.809 |
| Fetal beta | 1.216 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg03255775 | 5 | 98265241 | CHD1 | 5.67E-10 | -0.013 | 8.96E-7 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs17572651 | chr1 | 218943612 | CHD1 | 1105 | 0.15 | trans | ||
| rs16829545 | chr2 | 151977407 | CHD1 | 1105 | 1.736E-6 | trans | ||
| rs7584986 | chr2 | 184111432 | CHD1 | 1105 | 0.15 | trans | ||
| rs11139334 | chr9 | 84209393 | CHD1 | 1105 | 0.04 | trans | ||
| rs4112777 | chr13 | 99281457 | CHD1 | 1105 | 0.1 | trans | ||
| rs16955618 | chr15 | 29937543 | CHD1 | 1105 | 7.602E-7 | trans |
Section II. Transcriptome annotation
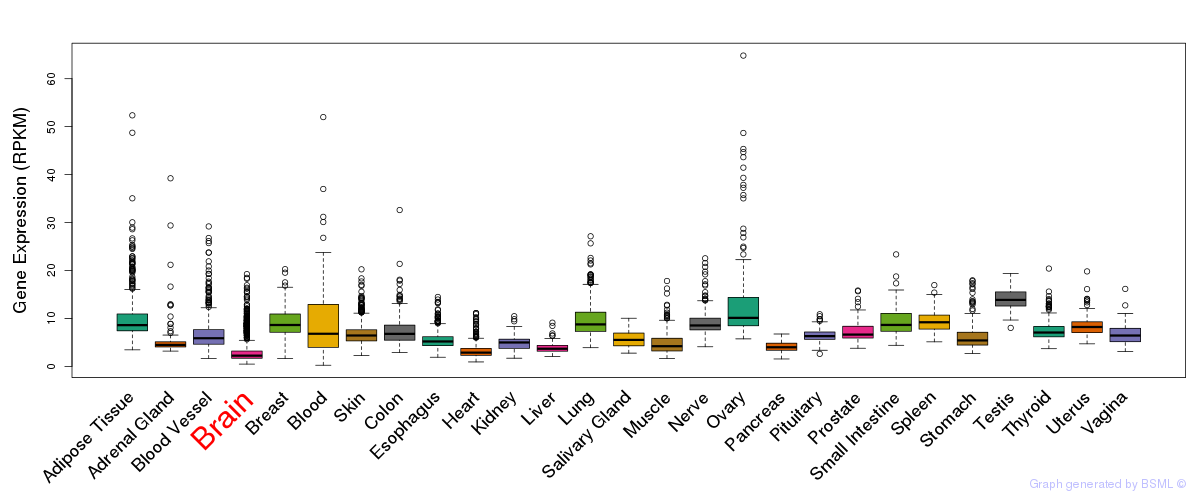
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| KLC2 | 0.85 | 0.87 |
| PANX2 | 0.84 | 0.78 |
| RAB3A | 0.79 | 0.84 |
| GRIN1 | 0.78 | 0.81 |
| KCNC3 | 0.78 | 0.80 |
| PDE4A | 0.78 | 0.81 |
| ATP1A1 | 0.78 | 0.80 |
| KCNJ11 | 0.77 | 0.77 |
| DCTN1 | 0.77 | 0.82 |
| AC021534.2 | 0.77 | 0.80 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.21 | -0.42 | -0.32 |
| AP002478.3 | -0.40 | -0.44 |
| RAB13 | -0.40 | -0.49 |
| GNG11 | -0.39 | -0.40 |
| IFI44 | -0.39 | -0.50 |
| AF347015.18 | -0.36 | -0.24 |
| C1orf54 | -0.36 | -0.38 |
| MYL12A | -0.36 | -0.44 |
| NOSTRIN | -0.36 | -0.35 |
| C1orf61 | -0.36 | -0.43 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PICCALUGA ANGIOIMMUNOBLASTIC LYMPHOMA DN | 136 | 86 | All SZGR 2.0 genes in this pathway |
| WATANABE RECTAL CANCER RADIOTHERAPY RESPONSIVE DN | 92 | 51 | All SZGR 2.0 genes in this pathway |
| CHOW RASSF1 TARGETS UP | 27 | 17 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| BERENJENO TRANSFORMED BY RHOA FOREVER DN | 31 | 23 | All SZGR 2.0 genes in this pathway |
| BERENJENO TRANSFORMED BY RHOA UP | 536 | 340 | All SZGR 2.0 genes in this pathway |
| HAMAI APOPTOSIS VIA TRAIL UP | 584 | 356 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 DN | 855 | 609 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 COMMON DN | 483 | 336 | All SZGR 2.0 genes in this pathway |
| FERREIRA EWINGS SARCOMA UNSTABLE VS STABLE UP | 167 | 92 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 6HR DN | 514 | 330 | All SZGR 2.0 genes in this pathway |
| PASQUALUCCI LYMPHOMA BY GC STAGE DN | 165 | 104 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL QTL TRANS | 882 | 572 | All SZGR 2.0 genes in this pathway |
| THEILGAARD NEUTROPHIL AT SKIN WOUND UP | 77 | 52 | All SZGR 2.0 genes in this pathway |
| BASSO CD40 SIGNALING UP | 101 | 76 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 16HR UP | 225 | 139 | All SZGR 2.0 genes in this pathway |
| RAMALHO STEMNESS UP | 206 | 118 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| DAZARD RESPONSE TO UV SCC DN | 123 | 86 | All SZGR 2.0 genes in this pathway |
| GENTILE UV RESPONSE CLUSTER D2 | 41 | 30 | All SZGR 2.0 genes in this pathway |
| GENTILE UV HIGH DOSE DN | 312 | 203 | All SZGR 2.0 genes in this pathway |
| DAZARD RESPONSE TO UV NHEK DN | 318 | 220 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 STIMULATED | 1022 | 619 | All SZGR 2.0 genes in this pathway |
| FOSTER TOLERANT MACROPHAGE DN | 409 | 268 | All SZGR 2.0 genes in this pathway |
| ACEVEDO NORMAL TISSUE ADJACENT TO LIVER TUMOR UP | 174 | 96 | All SZGR 2.0 genes in this pathway |
| BOCHKIS FOXA2 TARGETS | 425 | 261 | All SZGR 2.0 genes in this pathway |
| ZHANG TLX TARGETS 36HR DN | 185 | 116 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH INV 16 TRANSLOCATION | 422 | 277 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS DN | 882 | 538 | All SZGR 2.0 genes in this pathway |
| JUBAN TARGETS OF SPI1 AND FLI1 DN | 92 | 60 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG BOUND 8D | 658 | 397 | All SZGR 2.0 genes in this pathway |
| ZWANG CLASS 1 TRANSIENTLY INDUCED BY EGF | 516 | 308 | All SZGR 2.0 genes in this pathway |