Gene Page: TRIOBP
Summary ?
| GeneID | 11078 |
| Symbol | TRIOBP |
| Synonyms | DFNB28|HRIHFB2122|TAP68|TARA|dJ37E16.4 |
| Description | TRIO and F-actin binding protein |
| Reference | MIM:609761|HGNC:HGNC:17009|Ensembl:ENSG00000100106|HPRD:11028|Vega:OTTHUMG00000150657 |
| Gene type | protein-coding |
| Map location | 22q13.1 |
| Pascal p-value | 0.822 |
| Sherlock p-value | 0.48 |
| DEG p-value | DEG:Maycox_2009:CC_BA10_fold_change=1.13:CC_BA10_disease_P=0.0075:HBB_BA9_fold_change=1.19:HBB_BA9_disease_P=0.0292 |
| Fetal beta | 1.845 |
| eGene | Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| DEG:Maycox_2009 | Microarray to determine the expression of over 30000 mRNA transcripts in post-mortem tissue | We included 51 genes whose expression changes are common between two schizophrenia cohorts. | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
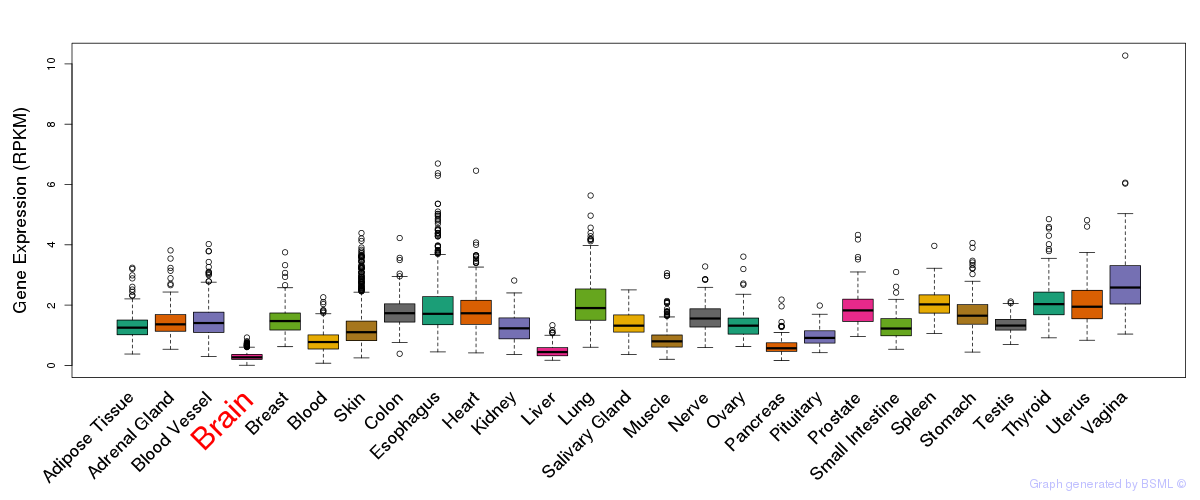
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MPDZ | 0.68 | 0.70 |
| GPR75 | 0.66 | 0.68 |
| ARHGEF10 | 0.65 | 0.69 |
| PIK3C2A | 0.65 | 0.64 |
| IQGAP1 | 0.64 | 0.65 |
| SLC41A1 | 0.64 | 0.67 |
| FAM120C | 0.64 | 0.62 |
| ZC3HAV1 | 0.64 | 0.63 |
| WWC3 | 0.63 | 0.67 |
| RREB1 | 0.63 | 0.62 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| RP11-3P17.1 | -0.33 | -0.36 |
| RPL31 | -0.31 | -0.32 |
| FAM159B | -0.31 | -0.31 |
| RPL35 | -0.31 | -0.35 |
| DDTL | -0.31 | -0.30 |
| PFDN5 | -0.30 | -0.30 |
| RPL27 | -0.30 | -0.32 |
| RPL13AP22 | -0.30 | -0.37 |
| AC087071.1 | -0.30 | -0.27 |
| GADD45GIP1 | -0.29 | -0.32 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PARENT MTOR SIGNALING UP | 567 | 375 | All SZGR 2.0 genes in this pathway |
| GAZDA DIAMOND BLACKFAN ANEMIA ERYTHROID DN | 493 | 298 | All SZGR 2.0 genes in this pathway |
| CHEMNITZ RESPONSE TO PROSTAGLANDIN E2 DN | 391 | 222 | All SZGR 2.0 genes in this pathway |
| SCIAN INVERSED TARGETS OF TP53 AND TP73 DN | 31 | 23 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 6HR DN | 514 | 330 | All SZGR 2.0 genes in this pathway |
| ROZANOV MMP14 TARGETS DN | 35 | 22 | All SZGR 2.0 genes in this pathway |
| NING CHRONIC OBSTRUCTIVE PULMONARY DISEASE UP | 157 | 105 | All SZGR 2.0 genes in this pathway |
| MOREAUX MULTIPLE MYELOMA BY TACI UP | 412 | 249 | All SZGR 2.0 genes in this pathway |
| MOREAUX B LYMPHOCYTE MATURATION BY TACI UP | 92 | 58 | All SZGR 2.0 genes in this pathway |
| LINDVALL IMMORTALIZED BY TERT UP | 78 | 48 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 18HR DN | 178 | 121 | All SZGR 2.0 genes in this pathway |
| BILD MYC ONCOGENIC SIGNATURE | 206 | 117 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR UP | 953 | 554 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR UP | 783 | 442 | All SZGR 2.0 genes in this pathway |
| RIGGI EWING SARCOMA PROGENITOR DN | 191 | 123 | All SZGR 2.0 genes in this pathway |
| TOOKER GEMCITABINE RESISTANCE DN | 122 | 84 | All SZGR 2.0 genes in this pathway |
| QI HYPOXIA | 140 | 96 | All SZGR 2.0 genes in this pathway |
| LU EZH2 TARGETS UP | 295 | 155 | All SZGR 2.0 genes in this pathway |
| FEVR CTNNB1 TARGETS UP | 682 | 433 | All SZGR 2.0 genes in this pathway |
| PURBEY TARGETS OF CTBP1 NOT SATB1 UP | 344 | 215 | All SZGR 2.0 genes in this pathway |
| KRIEG HYPOXIA NOT VIA KDM3A | 770 | 480 | All SZGR 2.0 genes in this pathway |
| FOSTER KDM1A TARGETS DN | 211 | 119 | All SZGR 2.0 genes in this pathway |