Gene Page: ADAMTS5
Summary ?
| GeneID | 11096 |
| Symbol | ADAMTS5 |
| Synonyms | ADAM-TS 11|ADAM-TS 5|ADAM-TS5|ADAMTS-11|ADAMTS-5|ADAMTS11|ADMP-2 |
| Description | ADAM metallopeptidase with thrombospondin type 1 motif 5 |
| Reference | MIM:605007|HGNC:HGNC:221|Ensembl:ENSG00000154736|HPRD:05420|Vega:OTTHUMG00000078686 |
| Gene type | protein-coding |
| Map location | 21q21.3 |
| Pascal p-value | 0.467 |
| Fetal beta | 0.669 |
| DMG | 1 (# studies) |
| eGene | Cerebellar Hemisphere Cerebellum Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg13601496 | 21 | 28339487 | ADAMTS5 | 5.3E-8 | -0.006 | 1.38E-5 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs6687849 | chr1 | 175904808 | ADAMTS5 | 11096 | 0 | trans | ||
| rs2481653 | chr1 | 176044120 | ADAMTS5 | 11096 | 0.01 | trans | ||
| rs2502827 | chr1 | 176044216 | ADAMTS5 | 11096 | 8.51E-6 | trans | ||
| rs12405921 | chr1 | 206695730 | ADAMTS5 | 11096 | 0.02 | trans | ||
| rs17572651 | chr1 | 218943612 | ADAMTS5 | 11096 | 0.01 | trans | ||
| rs882011 | chr2 | 22711668 | ADAMTS5 | 11096 | 0.12 | trans | ||
| rs16829545 | chr2 | 151977407 | ADAMTS5 | 11096 | 1.079E-7 | trans | ||
| rs3845734 | chr2 | 171125572 | ADAMTS5 | 11096 | 8.235E-5 | trans | ||
| rs7584986 | chr2 | 184111432 | ADAMTS5 | 11096 | 3.943E-12 | trans | ||
| rs6807632 | chr3 | 111395567 | ADAMTS5 | 11096 | 0 | trans | ||
| rs7681432 | chr4 | 56162058 | ADAMTS5 | 11096 | 0.04 | trans | ||
| rs7664496 | chr4 | 56252823 | ADAMTS5 | 11096 | 0.15 | trans | ||
| snp_a-2078569 | 0 | ADAMTS5 | 11096 | 0.03 | trans | |||
| rs17762315 | chr5 | 76807576 | ADAMTS5 | 11096 | 0.02 | trans | ||
| rs10491487 | chr5 | 80323367 | ADAMTS5 | 11096 | 0.01 | trans | ||
| rs1368303 | chr5 | 147672388 | ADAMTS5 | 11096 | 3.288E-6 | trans | ||
| rs4872540 | chr8 | 22598980 | ADAMTS5 | 11096 | 0.2 | trans | ||
| rs6989594 | chr8 | 126303866 | ADAMTS5 | 11096 | 0.07 | trans | ||
| rs11139334 | chr9 | 84209393 | ADAMTS5 | 11096 | 0.07 | trans | ||
| rs17263352 | chr9 | 124811572 | ADAMTS5 | 11096 | 0.02 | trans | ||
| rs457696 | chr13 | 32581313 | ADAMTS5 | 11096 | 0.18 | trans | ||
| rs7996604 | chr13 | 42224412 | ADAMTS5 | 11096 | 0.15 | trans | ||
| snp_a-1880548 | 0 | ADAMTS5 | 11096 | 0.14 | trans | |||
| rs1629937 | chr14 | 77059671 | ADAMTS5 | 11096 | 0.06 | trans | ||
| rs17104720 | chr14 | 77127308 | ADAMTS5 | 11096 | 1.159E-5 | trans | ||
| rs17104722 | chr14 | 77138109 | ADAMTS5 | 11096 | 0.18 | trans | ||
| rs17104782 | chr14 | 77156735 | ADAMTS5 | 11096 | 0.18 | trans | ||
| rs6574467 | chr14 | 79179744 | ADAMTS5 | 11096 | 0.17 | trans | ||
| rs10146003 | chr14 | 79191170 | ADAMTS5 | 11096 | 0.17 | trans | ||
| rs16955618 | chr15 | 29937543 | ADAMTS5 | 11096 | 1.216E-25 | trans | ||
| snp_a-1830894 | 0 | ADAMTS5 | 11096 | 0 | trans | |||
| rs3743068 | chr15 | 79047284 | ADAMTS5 | 11096 | 0.05 | trans | ||
| rs17823633 | chr17 | 68851458 | ADAMTS5 | 11096 | 0.04 | trans | ||
| rs17070739 | chr18 | 60819382 | ADAMTS5 | 11096 | 0.13 | trans | ||
| rs17085767 | chr18 | 69839397 | ADAMTS5 | 11096 | 0.05 | trans | ||
| rs749043 | chr20 | 46995700 | ADAMTS5 | 11096 | 0.02 | trans | ||
| rs5955025 | chrX | 142542997 | ADAMTS5 | 11096 | 0.18 | trans |
Section II. Transcriptome annotation
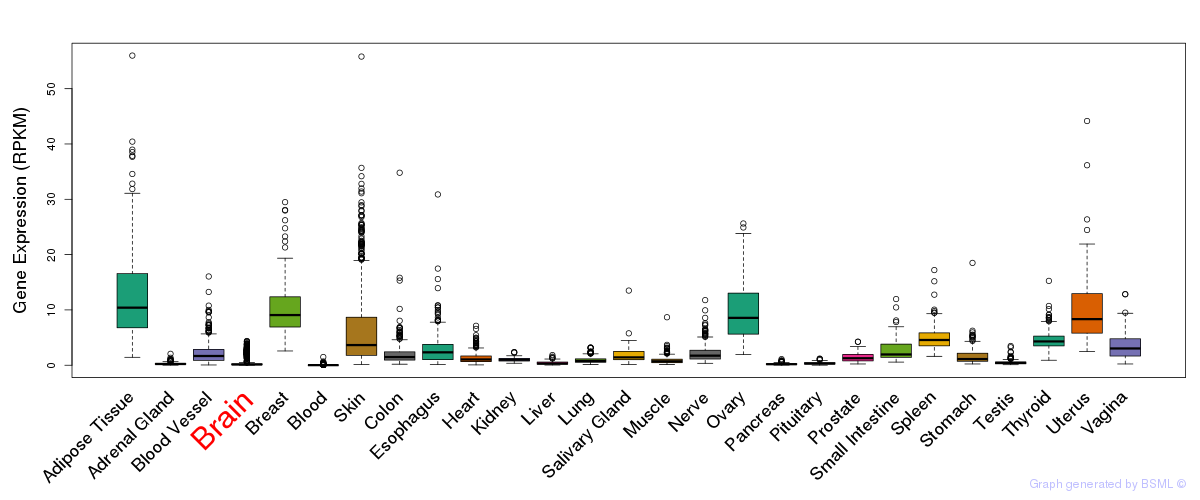
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| LIU PROSTATE CANCER DN | 481 | 290 | All SZGR 2.0 genes in this pathway |
| DAVICIONI TARGETS OF PAX FOXO1 FUSIONS UP | 255 | 177 | All SZGR 2.0 genes in this pathway |
| SENGUPTA NASOPHARYNGEAL CARCINOMA WITH LMP1 UP | 408 | 247 | All SZGR 2.0 genes in this pathway |
| TURASHVILI BREAST DUCTAL CARCINOMA VS DUCTAL NORMAL DN | 198 | 110 | All SZGR 2.0 genes in this pathway |
| TURASHVILI BREAST LOBULAR CARCINOMA VS LOBULAR NORMAL UP | 94 | 59 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS MESENCHYMAL DN | 460 | 312 | All SZGR 2.0 genes in this pathway |
| RODRIGUES NTN1 TARGETS DN | 158 | 102 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE LPS UP | 431 | 237 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION DN | 329 | 219 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| TANG SENESCENCE TP53 TARGETS UP | 33 | 20 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 4 5WK UP | 271 | 175 | All SZGR 2.0 genes in this pathway |
| BEGUM TARGETS OF PAX3 FOXO1 FUSION UP | 60 | 45 | All SZGR 2.0 genes in this pathway |
| WONG ENDMETRIUM CANCER DN | 82 | 53 | All SZGR 2.0 genes in this pathway |
| PEREZ TP53 TARGETS | 1174 | 695 | All SZGR 2.0 genes in this pathway |
| LIEN BREAST CARCINOMA METAPLASTIC | 35 | 25 | All SZGR 2.0 genes in this pathway |
| BUYTAERT PHOTODYNAMIC THERAPY STRESS UP | 811 | 508 | All SZGR 2.0 genes in this pathway |
| LIAO METASTASIS | 539 | 324 | All SZGR 2.0 genes in this pathway |
| BENPORATH SUZ12 TARGETS | 1038 | 678 | All SZGR 2.0 genes in this pathway |
| BENPORATH EED TARGETS | 1062 | 725 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS EARLY PROGENITOR | 532 | 309 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 DN | 830 | 547 | All SZGR 2.0 genes in this pathway |
| BILD HRAS ONCOGENIC SIGNATURE | 261 | 166 | All SZGR 2.0 genes in this pathway |
| HUPER BREAST BASAL VS LUMINAL DN | 59 | 44 | All SZGR 2.0 genes in this pathway |
| ZHANG TLX TARGETS 36HR DN | 185 | 116 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K4ME3 AND H3K27ME3 | 1069 | 729 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA CHEMOTAXIS DN | 457 | 302 | All SZGR 2.0 genes in this pathway |
| MEISSNER NPC HCP WITH H3K4ME2 | 491 | 319 | All SZGR 2.0 genes in this pathway |
| HOELZEL NF1 TARGETS DN | 115 | 73 | All SZGR 2.0 genes in this pathway |
| MIYAGAWA TARGETS OF EWSR1 ETS FUSIONS DN | 229 | 135 | All SZGR 2.0 genes in this pathway |
| KATSANOU ELAVL1 TARGETS UP | 169 | 105 | All SZGR 2.0 genes in this pathway |
| SERVITJA LIVER HNF1A TARGETS UP | 135 | 96 | All SZGR 2.0 genes in this pathway |
| PLASARI TGFB1 TARGETS 10HR DN | 244 | 157 | All SZGR 2.0 genes in this pathway |
| NABA ECM REGULATORS | 238 | 125 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME ASSOCIATED | 753 | 411 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME | 1028 | 559 | All SZGR 2.0 genes in this pathway |