Gene Page: RASSF1
Summary ?
| GeneID | 11186 |
| Symbol | RASSF1 |
| Synonyms | 123F2|NORE2A|RASSF1A|RDA32|REH3P21 |
| Description | Ras association domain family member 1 |
| Reference | MIM:605082|HGNC:HGNC:9882|Ensembl:ENSG00000068028|HPRD:05470|Vega:OTTHUMG00000149958 |
| Gene type | protein-coding |
| Map location | 3p21.3 |
| Pascal p-value | 0.122 |
| Sherlock p-value | 0.355 |
| Fetal beta | 1.023 |
| DMG | 1 (# studies) |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:vanEijk_2014 | Genome-wide DNA methylation analysis | This dataset includes 432 differentially methylated CpG sites corresponding to 391 unique transcripts between schizophrenia patients (n=260) and unaffected controls (n=250). | 3 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg14358502 | 3 | 50337045 | RASSF1 | 4.23E-5 | 5.175 | DMG:vanEijk_2014 | |
| cg15877314 | 3 | 50273895 | RASSF1 | 0.001 | 5.147 | DMG:vanEijk_2014 | |
| cg17272094 | 3 | 50317223 | RASSF1 | 9.178E-4 | -6.907 | DMG:vanEijk_2014 |
Section II. Transcriptome annotation
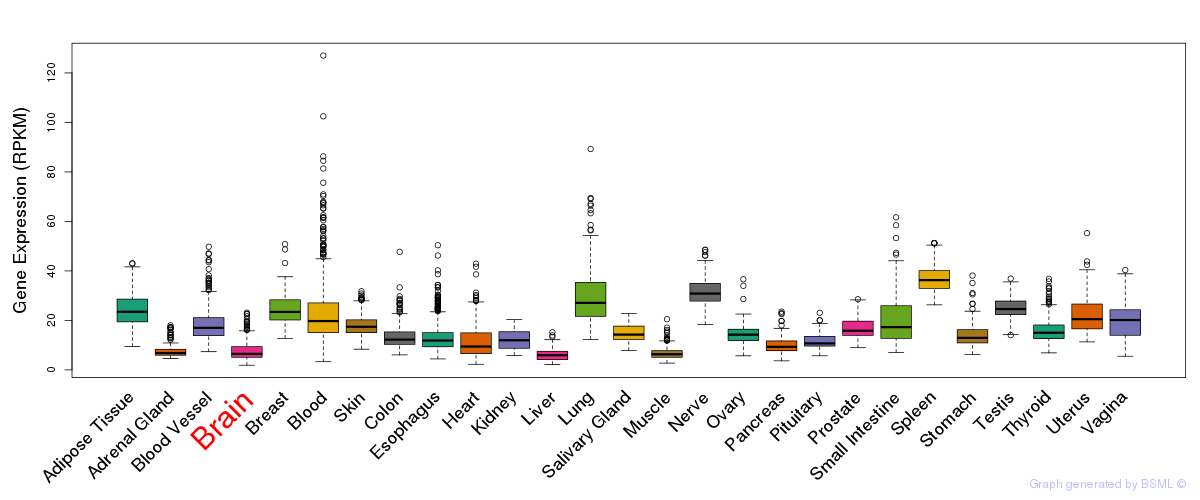
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ATP2B4 | ATP2B2 | DKFZp686G08106 | DKFZp686M088 | MXRA1 | PMCA4 | PMCA4b | PMCA4x | ATPase, Ca++ transporting, plasma membrane 4 | PMCA4b interacts with RASSF1A. | BIND | 15145946 |
| ATP2B4 | ATP2B2 | DKFZp686G08106 | DKFZp686M088 | MXRA1 | PMCA4 | PMCA4b | PMCA4x | ATPase, Ca++ transporting, plasma membrane 4 | PMCA4b interacts with RASSF1C. | BIND | 15145946 |
| ATP2B4 | ATP2B2 | DKFZp686G08106 | DKFZp686M088 | MXRA1 | PMCA4 | PMCA4b | PMCA4x | ATPase, Ca++ transporting, plasma membrane 4 | PMCA4b interacts with RASSF1E. | BIND | 15145946 |
| CNKSR1 | CNK | CNK1 | KSR | connector enhancer of kinase suppressor of Ras 1 | - | HPRD,BioGRID | 15075335 |
| EXOSC8 | CIP3 | EAP2 | OIP2 | RP11-421P11.3 | RRP43 | Rrp43p | bA421P11.3 | p9 | exosome component 8 | - | HPRD,BioGRID | 15231747 |
| HRAS | C-BAS/HAS | C-H-RAS | C-HA-RAS1 | CTLO | H-RASIDX | HAMSV | HRAS1 | K-RAS | N-RAS | RASH1 | v-Ha-ras Harvey rat sarcoma viral oncogene homolog | - | HPRD,BioGRID | 10998413 |
| MAP1B | DKFZp686E1099 | DKFZp686F1345 | FLJ38954 | FUTSCH | MAP5 | microtubule-associated protein 1B | Affinity Capture-Western Two-hybrid | BioGRID | 15205320 |
| MAP1S | BPY2IP1 | C19orf5 | FLJ10669 | MAP8 | MGC133087 | VCY2IP-1 | VCY2IP1 | microtubule-associated protein 1S | Affinity Capture-Western Two-hybrid | BioGRID | 12762840 |15205320 |15753381 |
| MOAP1 | MAP-1 | PNMA4 | modulator of apoptosis 1 | MAP-1 interacts with RASSF1A. | BIND | 15949439 |
| RASSF1 | 123F2 | NORE2A | RASSF1A | RDA32 | REH3P21 | Ras association (RalGDS/AF-6) domain family member 1 | - | HPRD,BioGRID | 11857081 |
| RASSF5 | MGC10823 | MGC17344 | Maxp1 | NORE1 | NORE1A | NORE1B | RAPL | RASSF3 | Ras association (RalGDS/AF-6) domain family member 5 | - | HPRD,BioGRID | 11857081 |
| TNFRSF10A | APO2 | CD261 | DR4 | MGC9365 | TRAILR-1 | TRAILR1 | tumor necrosis factor receptor superfamily, member 10a | TRAIL-R1 interacts with RASSF1A. | BIND | 15949439 |
| TNFRSF1A | CD120a | FPF | MGC19588 | TBP1 | TNF-R | TNF-R-I | TNF-R55 | TNFAR | TNFR1 | TNFR55 | TNFR60 | p55 | p55-R | p60 | tumor necrosis factor receptor superfamily, member 1A | TNF-R1 interacts with RASSF1A. | BIND | 15949439 |
| TUBA4A | FLJ30169 | H2-ALPHA | TUBA1 | tubulin, alpha 4a | Affinity Capture-Western | BioGRID | 15205320 |
| TUBB | M40 | MGC117247 | MGC16435 | OK/SW-cl.56 | TUBB1 | TUBB5 | tubulin, beta | Reconstituted Complex | BioGRID | 14603253 |
| TUBB2A | TUBB | TUBB2 | dJ40E16.7 | tubulin, beta 2A | - | HPRD | 14603253 |
| TUBG1 | TUBG | TUBGCP1 | tubulin, gamma 1 | - | HPRD,BioGRID | 14603253 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG PATHWAYS IN CANCER | 328 | 259 | All SZGR 2.0 genes in this pathway |
| KEGG BLADDER CANCER | 42 | 33 | All SZGR 2.0 genes in this pathway |
| KEGG NON SMALL CELL LUNG CANCER | 54 | 47 | All SZGR 2.0 genes in this pathway |
| PID P53 REGULATION PATHWAY | 59 | 50 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| UDAYAKUMAR MED1 TARGETS DN | 240 | 171 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND DOXORUBICIN DN | 770 | 415 | All SZGR 2.0 genes in this pathway |
| HESSON TUMOR SUPPRESSOR CLUSTER 3P21 3 | 7 | 5 | All SZGR 2.0 genes in this pathway |
| JAZAG TGFB1 SIGNALING VIA SMAD4 DN | 66 | 38 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| OHM METHYLATED IN ADULT CANCERS | 27 | 23 | All SZGR 2.0 genes in this pathway |
| BUYTAERT PHOTODYNAMIC THERAPY STRESS UP | 811 | 508 | All SZGR 2.0 genes in this pathway |
| GARCIA TARGETS OF FLI1 AND DAX1 DN | 176 | 104 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC MAX TARGETS | 775 | 494 | All SZGR 2.0 genes in this pathway |
| WANG TARGETS OF MLL CBP FUSION DN | 45 | 31 | All SZGR 2.0 genes in this pathway |
| MEDINA SMARCA4 TARGETS | 44 | 29 | All SZGR 2.0 genes in this pathway |
| HOQUE METHYLATED IN CANCER | 56 | 45 | All SZGR 2.0 genes in this pathway |
| YAUCH HEDGEHOG SIGNALING PARACRINE DN | 264 | 159 | All SZGR 2.0 genes in this pathway |
| CHEN METABOLIC SYNDROM NETWORK | 1210 | 725 | All SZGR 2.0 genes in this pathway |
| LI PROSTATE CANCER EPIGENETIC | 30 | 22 | All SZGR 2.0 genes in this pathway |
| MARTENS TRETINOIN RESPONSE DN | 841 | 431 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP B | 549 | 316 | All SZGR 2.0 genes in this pathway |
| KRIEG HYPOXIA NOT VIA KDM3A | 770 | 480 | All SZGR 2.0 genes in this pathway |
| FOSTER KDM1A TARGETS DN | 211 | 119 | All SZGR 2.0 genes in this pathway |