Gene Page: CHEK2
Summary ?
| GeneID | 11200 |
| Symbol | CHEK2 |
| Synonyms | CDS1|CHK2|HuCds1|LFS2|PP1425|RAD53|hCds1 |
| Description | checkpoint kinase 2 |
| Reference | MIM:604373|HGNC:HGNC:16627|Ensembl:ENSG00000183765|HPRD:05084|Vega:OTTHUMG00000151023 |
| Gene type | protein-coding |
| Map location | 22q12.1 |
| Pascal p-value | 0.004 |
| Sherlock p-value | 0.039 |
| Fetal beta | 1.082 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
| DNM:Gulsuner_2013 | Whole Exome Sequencing analysis | 155 DNMs identified by exome sequencing of quads or trios of schizophrenia individuals and their parents. | |
| GSMA_I | Genome scan meta-analysis | Psr: 0.031 |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| CHEK2 | chr22 | 29121000 | T | C | NM_001005735 NM_001257387 NM_007194 NM_145862 | p.229N>S . p.186N>S p.186N>S | missense 5-UTR missense missense | Schizophrenia | DNM:Gulsuner_2013 |
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg13900031 | 22 | 29137598 | CHEK2 | 7.58E-9 | -0.019 | 3.73E-6 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs1578978 | chr8 | 118196802 | CHEK2 | 11200 | 0.19 | trans |
Section II. Transcriptome annotation
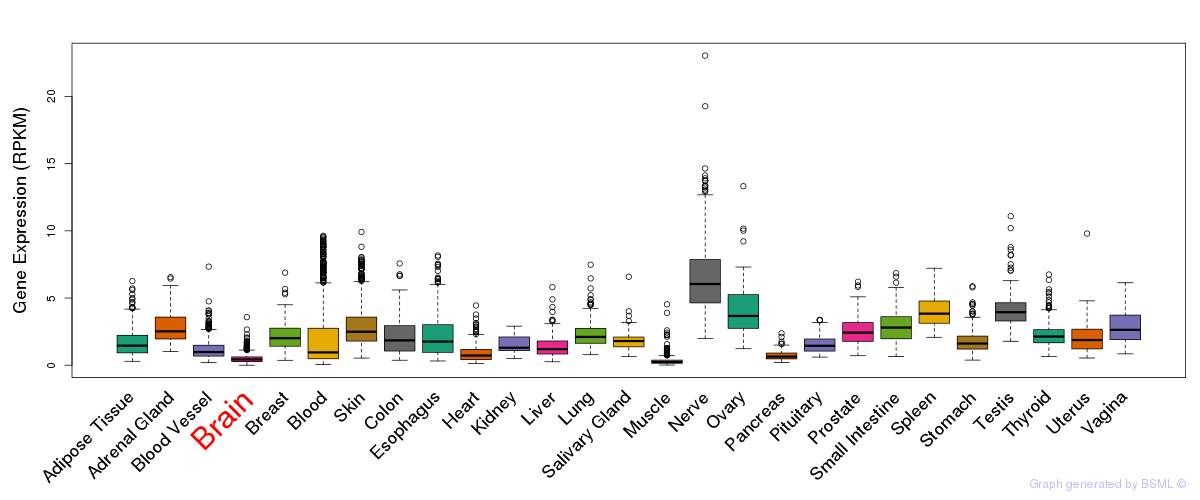
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0000287 | magnesium ion binding | IEA | - | |
| GO:0005515 | protein binding | IPI | 12402044 | |
| GO:0005524 | ATP binding | IEA | - | |
| GO:0004674 | protein serine/threonine kinase activity | IEA | - | |
| GO:0004674 | protein serine/threonine kinase activity | TAS | 9836640 | |
| GO:0016740 | transferase activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0000077 | DNA damage checkpoint | TAS | 9889122 |10617473 | |
| GO:0006468 | protein amino acid phosphorylation | IEA | - | |
| GO:0008630 | DNA damage response, signal transduction resulting in induction of apoptosis | IDA | 12402044 | |
| GO:0007049 | cell cycle | IEA | - | |
| GO:0010332 | response to gamma radiation | IEA | - | |
| GO:0042770 | DNA damage response, signal transduction | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005634 | nucleus | IEA | - | |
| GO:0016605 | PML body | IDA | 12402044 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ATM | AT1 | ATA | ATC | ATD | ATDC | ATE | DKFZp781A0353 | MGC74674 | TEL1 | TELO1 | ataxia telangiectasia mutated | ATM phosphorylates Chk2 at threonine 68. | BIND | 15790808 |
| ATM | AT1 | ATA | ATC | ATD | ATDC | ATE | DKFZp781A0353 | MGC74674 | TEL1 | TELO1 | ataxia telangiectasia mutated | ATM phosphorylates Chk2 at threonine 68. | BIND | 15064416 |
| ATMIN | ASCIZ | DKFZp779K1455 | FLJ76795 | KIAA0431 | ZNF822 | ATM interactor | ASCIZ interacts with an unspecified isoform of CHK2. | BIND | 15933716 |
| BRCA1 | BRCAI | BRCC1 | IRIS | PSCP | RNF53 | breast cancer 1, early onset | - | HPRD,BioGRID | 10724175 |
| BRCA2 | BRCC2 | FACD | FAD | FAD1 | FANCB | FANCD | FANCD1 | breast cancer 2, early onset | Affinity Capture-Western | BioGRID | 17525332 |
| DBF4 | ASK | CHIF | DBF4A | ZDBF1 | DBF4 homolog (S. cerevisiae) | - | HPRD,BioGRID | 12441400 |
| ERCC6 | ARMD5 | CKN2 | COFS | COFS1 | CSB | RAD26 | excision repair cross-complementing rodent repair deficiency, complementation group 6 | - | HPRD | 9450932 |
| GINS2 | HSPC037 | PSF2 | Pfs2 | GINS complex subunit 2 (Psf2 homolog) | Affinity Capture-Western | BioGRID | 17525332 |
| MDC1 | DKFZp781A0122 | KIAA0170 | MGC166888 | NFBD1 | mediator of DNA damage checkpoint 1 | - | HPRD,BioGRID | 12551934 |12607004 |
| MRC1 | CD206 | CLEC13D | MMR | mannose receptor, C type 1 | - | HPRD | 11715017 |
| MSH2 | COCA1 | FCC1 | HNPCC | HNPCC1 | LCFS2 | mutS homolog 2, colon cancer, nonpolyposis type 1 (E. coli) | - | HPRD,BioGRID | 12447371 |
| MUS81 | FLJ21012 | FLJ44872 | MUS81 endonuclease homolog (S. cerevisiae) | - | HPRD,BioGRID | 11741546 |
| PLK1 | PLK | STPK13 | polo-like kinase 1 (Drosophila) | - | HPRD,BioGRID | 12493754 |
| PLK3 | CNK | FNK | PRK | polo-like kinase 3 (Drosophila) | - | HPRD,BioGRID | 12242661 |
| RAD50 | RAD50-2 | hRad50 | RAD50 homolog (S. cerevisiae) | Affinity Capture-Western | BioGRID | 17525332 |
| RAD9A | RAD9 | RAD9 homolog A (S. pombe) | - | HPRD | 12049741 |
| TP53BP1 | 53BP1 | FLJ41424 | MGC138366 | p202 | tumor protein p53 binding protein 1 | Affinity Capture-Western | BioGRID | 17525332 |
| TSSK1B | FKSG81 | SPOGA4 | STK22D | TSSK1 | testis-specific serine kinase 1B | - | HPRD | 12556502 |
| VCP | IBMPFD | MGC131997 | MGC148092 | MGC8560 | TERA | p97 | valosin-containing protein | Affinity Capture-Western | BioGRID | 17525332 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG CELL CYCLE | 128 | 84 | All SZGR 2.0 genes in this pathway |
| KEGG P53 SIGNALING PATHWAY | 69 | 45 | All SZGR 2.0 genes in this pathway |
| BIOCARTA ATM PATHWAY | 20 | 16 | All SZGR 2.0 genes in this pathway |
| BIOCARTA G2 PATHWAY | 24 | 20 | All SZGR 2.0 genes in this pathway |
| BIOCARTA ATRBRCA PATHWAY | 21 | 12 | All SZGR 2.0 genes in this pathway |
| PID ATM PATHWAY | 34 | 25 | All SZGR 2.0 genes in this pathway |
| PID FOXM1 PATHWAY | 40 | 30 | All SZGR 2.0 genes in this pathway |
| PID P53 REGULATION PATHWAY | 59 | 50 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CYCLE | 421 | 253 | All SZGR 2.0 genes in this pathway |
| REACTOME P53 INDEPENDENT G1 S DNA DAMAGE CHECKPOINT | 51 | 31 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CYCLE CHECKPOINTS | 124 | 70 | All SZGR 2.0 genes in this pathway |
| REACTOME G2 M CHECKPOINTS | 45 | 23 | All SZGR 2.0 genes in this pathway |
| REACTOME G2 M DNA DAMAGE CHECKPOINT | 12 | 8 | All SZGR 2.0 genes in this pathway |
| LIU SOX4 TARGETS DN | 309 | 191 | All SZGR 2.0 genes in this pathway |
| CASORELLI ACUTE PROMYELOCYTIC LEUKEMIA DN | 663 | 425 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION UP | 1278 | 748 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA DN | 1375 | 806 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| WAKASUGI HAVE ZNF143 BINDING SITES | 58 | 33 | All SZGR 2.0 genes in this pathway |
| PUJANA BREAST CANCER LIT INT NETWORK | 101 | 73 | All SZGR 2.0 genes in this pathway |
| PUJANA XPRSS INT NETWORK | 168 | 103 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA2 PCC NETWORK | 423 | 265 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| PUJANA CHEK2 PCC NETWORK | 779 | 480 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA CENTERED NETWORK | 117 | 72 | All SZGR 2.0 genes in this pathway |
| KAUFFMANN MELANOMA RELAPSE UP | 61 | 25 | All SZGR 2.0 genes in this pathway |
| GROSS HYPOXIA VIA ELK3 ONLY DN | 44 | 30 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES 1 | 379 | 235 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES 2 | 40 | 21 | All SZGR 2.0 genes in this pathway |
| BENPORATH CYCLING GENES | 648 | 385 | All SZGR 2.0 genes in this pathway |
| KAUFFMANN DNA REPAIR GENES | 230 | 137 | All SZGR 2.0 genes in this pathway |
| GOLDRATH HOMEOSTATIC PROLIFERATION | 171 | 102 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 14HR DN | 298 | 200 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 48HR DN | 504 | 323 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 12HR UP | 111 | 68 | All SZGR 2.0 genes in this pathway |
| WANG CISPLATIN RESPONSE AND XPC UP | 202 | 115 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY E2F4 UNSTIMULATED | 728 | 415 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 TARGETS DN | 543 | 317 | All SZGR 2.0 genes in this pathway |
| MARTINEZ TP53 TARGETS DN | 593 | 372 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 AND TP53 TARGETS UP | 601 | 369 | All SZGR 2.0 genes in this pathway |
| MUELLER PLURINET | 299 | 189 | All SZGR 2.0 genes in this pathway |
| FIRESTEIN CTNNB1 PATHWAY | 33 | 23 | All SZGR 2.0 genes in this pathway |
| GRESHOCK CANCER COPY NUMBER UP | 323 | 240 | All SZGR 2.0 genes in this pathway |
| AGUIRRE PANCREATIC CANCER COPY NUMBER DN | 238 | 145 | All SZGR 2.0 genes in this pathway |
| CROONQUIST NRAS SIGNALING DN | 72 | 47 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA CLASSES UP | 605 | 377 | All SZGR 2.0 genes in this pathway |
| WONG EMBRYONIC STEM CELL CORE | 335 | 193 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 4HR DN | 254 | 158 | All SZGR 2.0 genes in this pathway |
| WHITFIELD CELL CYCLE G2 | 182 | 102 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE LATE | 1137 | 655 | All SZGR 2.0 genes in this pathway |
| FEVR CTNNB1 TARGETS DN | 553 | 343 | All SZGR 2.0 genes in this pathway |
| PURBEY TARGETS OF CTBP1 AND SATB1 UP | 87 | 52 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION UP | 570 | 339 | All SZGR 2.0 genes in this pathway |