| KEGG APOPTOSIS
| 88 | 62 | All SZGR 2.0 genes in this pathway |
| KEGG NEUROTROPHIN SIGNALING PATHWAY
| 126 | 103 | All SZGR 2.0 genes in this pathway |
| BIOCARTA IL1R PATHWAY
| 33 | 29 | All SZGR 2.0 genes in this pathway |
| PID IL1 PATHWAY
| 34 | 28 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY ILS
| 107 | 86 | All SZGR 2.0 genes in this pathway |
| REACTOME IL1 SIGNALING
| 39 | 30 | All SZGR 2.0 genes in this pathway |
| REACTOME MYD88 MAL CASCADE INITIATED ON PLASMA MEMBRANE
| 83 | 63 | All SZGR 2.0 genes in this pathway |
| REACTOME INNATE IMMUNE SYSTEM
| 279 | 178 | All SZGR 2.0 genes in this pathway |
| REACTOME ACTIVATED TLR4 SIGNALLING
| 93 | 69 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM
| 933 | 616 | All SZGR 2.0 genes in this pathway |
| REACTOME TOLL RECEPTOR CASCADES
| 118 | 84 | All SZGR 2.0 genes in this pathway |
| REACTOME CYTOKINE SIGNALING IN IMMUNE SYSTEM
| 270 | 204 | All SZGR 2.0 genes in this pathway |
| PICCALUGA ANGIOIMMUNOBLASTIC LYMPHOMA UP
| 205 | 140 | All SZGR 2.0 genes in this pathway |
| SCHUETZ BREAST CANCER DUCTAL INVASIVE UP
| 351 | 230 | All SZGR 2.0 genes in this pathway |
| SENGUPTA NASOPHARYNGEAL CARCINOMA WITH LMP1 UP
| 408 | 247 | All SZGR 2.0 genes in this pathway |
| SMIRNOV CIRCULATING ENDOTHELIOCYTES IN CANCER UP
| 158 | 103 | All SZGR 2.0 genes in this pathway |
| OSMAN BLADDER CANCER UP
| 404 | 246 | All SZGR 2.0 genes in this pathway |
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION ERYTHROCYTE UP
| 157 | 104 | All SZGR 2.0 genes in this pathway |
| HEIDENBLAD AMPLICON 8Q24 DN
| 46 | 28 | All SZGR 2.0 genes in this pathway |
| LOPEZ MBD TARGETS
| 957 | 597 | All SZGR 2.0 genes in this pathway |
| BENPORATH EED TARGETS
| 1062 | 725 | All SZGR 2.0 genes in this pathway |
| FOSTER TOLERANT MACROPHAGE UP
| 156 | 92 | All SZGR 2.0 genes in this pathway |
| BONOME OVARIAN CANCER SURVIVAL SUBOPTIMAL DEBULKING
| 510 | 309 | All SZGR 2.0 genes in this pathway |
| CHUNG BLISTER CYTOTOXICITY DN
| 44 | 29 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO CSF2RB AND IL4 DN
| 315 | 201 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF DN
| 235 | 144 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF VS CSF2RB AND IL4 UP
| 408 | 276 | All SZGR 2.0 genes in this pathway |
| CHANG CORE SERUM RESPONSE DN
| 209 | 137 | All SZGR 2.0 genes in this pathway |
| SHEDDEN LUNG CANCER GOOD SURVIVAL A12
| 317 | 177 | All SZGR 2.0 genes in this pathway |
| MEISSNER NPC HCP WITH H3 UNMETHYLATED
| 536 | 296 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K4ME3 AND H3K27ME3
| 1069 | 729 | All SZGR 2.0 genes in this pathway |
| MARTENS TRETINOIN RESPONSE DN
| 841 | 431 | All SZGR 2.0 genes in this pathway |
| FIGUEROA AML METHYLATION CLUSTER 6 UP
| 140 | 81 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS DN
| 882 | 538 | All SZGR 2.0 genes in this pathway |
| HUANG GATA2 TARGETS UP
| 149 | 96 | All SZGR 2.0 genes in this pathway |
| GHANDHI BYSTANDER IRRADIATION UP
| 86 | 54 | All SZGR 2.0 genes in this pathway |
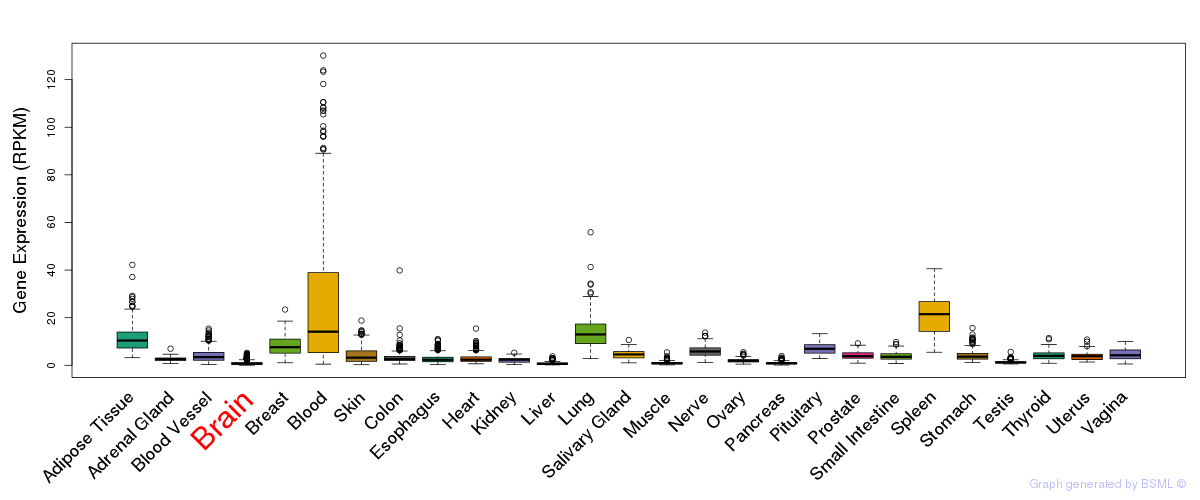
 Differentially methylated gene
Differentially methylated gene eQTL annotation
eQTL annotation