Gene Page: DDX20
Summary ?
| GeneID | 11218 |
| Symbol | DDX20 |
| Synonyms | DP103|GEMIN3 |
| Description | DEAD-box helicase 20 |
| Reference | MIM:606168|HGNC:HGNC:2743|Ensembl:ENSG00000064703|HPRD:05859|Vega:OTTHUMG00000011956 |
| Gene type | protein-coding |
| Map location | 1p21.1-p13.2 |
| Pascal p-value | 0.129 |
| Fetal beta | 0.888 |
| eGene | Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
| DNM:Guipponi_2014 | Whole Exome Sequencing analysis | 49 DNMs were identified by comparing the exome of 53 individuals with sporadic SCZ and of their non-affected parents | |
| GSMA_I | Genome scan meta-analysis | Psr: 0.0235 |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| DDX20 | G | C | NM_007204 | p.D55H | missense | 0 | 1 | Schizophrenia | DNM:Guipponi_2014 |
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg17838705 | 1 | 112298602 | DDX20;C1orf183 | 1.978E-4 | -0.218 | 0.035 | DMG:Wockner_2014 |
Section II. Transcriptome annotation
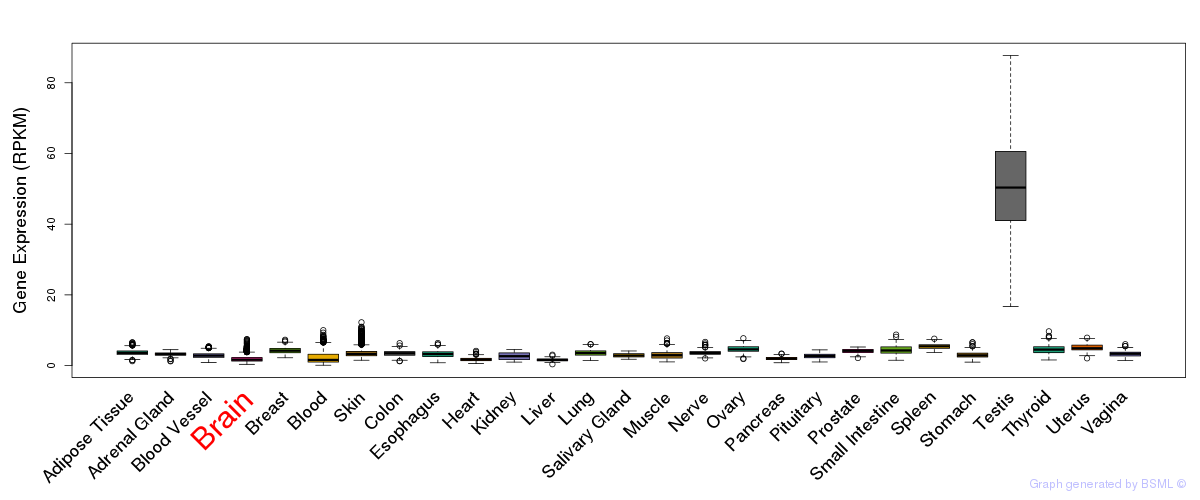
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0003677 | DNA binding | IEA | - | |
| GO:0004004 | ATP-dependent RNA helicase activity | TAS | 10383418 | |
| GO:0005515 | protein binding | IPI | 11714716 | |
| GO:0005515 | protein binding | ISS | - | |
| GO:0005524 | ATP binding | IEA | - | |
| GO:0016787 | hydrolase activity | IEA | - | |
| GO:0016564 | transcription repressor activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0000122 | negative regulation of transcription from RNA polymerase II promoter | IEA | - | |
| GO:0000244 | assembly of spliceosomal tri-snRNP | TAS | 10601333 | |
| GO:0006397 | mRNA processing | IEA | - | |
| GO:0008380 | RNA splicing | IEA | - | |
| GO:0006917 | induction of apoptosis | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005829 | cytosol | EXP | 10531003 | |
| GO:0005856 | cytoskeleton | TAS | 10383418 | |
| GO:0005634 | nucleus | TAS | 10383418 | |
| GO:0005654 | nucleoplasm | EXP | 10531003 | |
| GO:0005681 | spliceosome | IEA | - | |
| GO:0005737 | cytoplasm | TAS | 10601333 | |
| GO:0017053 | transcriptional repressor complex | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| BAT2 | D6S51 | D6S51E | DKFZp686D09175 | G2 | HLA-B associated transcript 2 | - | HPRD,BioGRID | 14667819 |
| DHX9 | DDX9 | LKP | NDHII | RHA | DEAH (Asp-Glu-Ala-His) box polypeptide 9 | Affinity Capture-Western | BioGRID | 11149922 |
| EIF2C2 | AGO2 | MGC3183 | Q10 | eukaryotic translation initiation factor 2C, 2 | - | HPRD,BioGRID | 11914277 |14970384 |
| FBL | FIB | FLRN | RNU3IP1 | fibrillarin | Affinity Capture-Western | BioGRID | 11509230 |
| GAR1 | NOLA1 | GAR1 ribonucleoprotein homolog (yeast) | Affinity Capture-Western | BioGRID | 11509230 |
| GEMIN4 | DKFZp434B131 | DKFZp434D174 | HC56 | HCAP1 | HHRF-1 | p97 | gem (nuclear organelle) associated protein 4 | Affinity Capture-Western Reconstituted Complex | BioGRID | 10725331 |11914277 |12668731 |
| GEMIN5 | DKFZp586M1824 | MGC142174 | gem (nuclear organelle) associated protein 5 | - | HPRD,BioGRID | 11714716 |
| GEMIN6 | FLJ23459 | gem (nuclear organelle) associated protein 6 | Affinity Capture-Western | BioGRID | 11748230 |
| GSK3B | - | glycogen synthase kinase 3 beta | Affinity Capture-MS | BioGRID | 17353931 |
| LSM2 | C6orf28 | G7b | YBL026W | snRNP | LSM2 homolog, U6 small nuclear RNA associated (S. cerevisiae) | Affinity Capture-Western Reconstituted Complex | BioGRID | 10601333 |
| PHB2 | BAP | BCAP37 | Bap37 | MGC117268 | PNAS-141 | REA | p22 | prohibitin 2 | Affinity Capture-MS | BioGRID | 17353931 |
| PNKP | PNK | polynucleotide kinase 3'-phosphatase | Affinity Capture-MS | BioGRID | 17353931 |
| POLR1E | FLJ13390 | FLJ13970 | PAF53 | PRAF1 | RP11-405L18.3 | polymerase (RNA) I polypeptide E, 53kDa | Affinity Capture-MS | BioGRID | 17353931 |
| PPP4R2 | MGC131930 | protein phosphatase 4, regulatory subunit 2 | Affinity Capture-MS Affinity Capture-Western | BioGRID | 12668731 |
| RNPS1 | E5.1 | MGC117332 | RNA binding protein S1, serine-rich domain | Affinity Capture-MS | BioGRID | 17353931 |
| SIP1 | GEMIN2 | SIP1-delta | survival of motor neuron protein interacting protein 1 | Affinity Capture-Western Reconstituted Complex | BioGRID | 10601333 |11914277 |
| SMN1 | BCD541 | SMA | SMA1 | SMA2 | SMA3 | SMA4 | SMA@ | SMN | SMNT | T-BCD541 | survival of motor neuron 1, telomeric | - | HPRD,BioGRID | 10601333 |
| SMN2 | BCD541 | C-BCD541 | FLJ76644 | MGC20996 | MGC5208 | SMNC | survival of motor neuron 2, centromeric | - | HPRD | 10942426 |
| SNRPB | COD | SNRPB1 | SmB/SmB' | snRNP-B | small nuclear ribonucleoprotein polypeptides B and B1 | - | HPRD,BioGRID | 10601333 |
| SNRPD1 | HsT2456 | SMD1 | SNRPD | small nuclear ribonucleoprotein D1 polypeptide 16kDa | - | HPRD,BioGRID | 10601333 |
| SNRPD2 | SMD2 | SNRPD1 | small nuclear ribonucleoprotein D2 polypeptide 16.5kDa | - | HPRD,BioGRID | 10601333 |
| SNRPD3 | SMD3 | small nuclear ribonucleoprotein D3 polypeptide 18kDa | - | HPRD,BioGRID | 10601333 |
| SNRPE | B-raf | SME | small nuclear ribonucleoprotein polypeptide E | - | HPRD,BioGRID | 10601333 |
| SNRPF | SMF | small nuclear ribonucleoprotein polypeptide F | - | HPRD,BioGRID | 10601333 |
| SNRPG | MGC117317 | SMG | small nuclear ribonucleoprotein polypeptide G | - | HPRD,BioGRID | 10601333 |
| SNUPN | KPNBL | RNUT1 | Snurportin1 | snurportin 1 | Reconstituted Complex | BioGRID | 12095920 |
| STXBP3 | MUNC18-3 | MUNC18C | PSP | UNC-18C | syntaxin binding protein 3 | - | HPRD | 12065586 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| BIOCARTA ETS PATHWAY | 18 | 12 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF NON CODING RNA | 49 | 29 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF RNA | 330 | 155 | All SZGR 2.0 genes in this pathway |
| GARY CD5 TARGETS DN | 431 | 263 | All SZGR 2.0 genes in this pathway |
| WANG LMO4 TARGETS DN | 352 | 225 | All SZGR 2.0 genes in this pathway |
| WEI MYCN TARGETS WITH E BOX | 795 | 478 | All SZGR 2.0 genes in this pathway |
| BENPORATH NANOG TARGETS | 988 | 594 | All SZGR 2.0 genes in this pathway |
| SAKAI TUMOR INFILTRATING MONOCYTES DN | 81 | 51 | All SZGR 2.0 genes in this pathway |
| SHEPARD CRUSH AND BURN MUTANT DN | 185 | 111 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS INTERMEDIATE PROGENITOR | 149 | 84 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RESPONSE TO TRABECTEDIN DN | 271 | 175 | All SZGR 2.0 genes in this pathway |
| ZHANG TLX TARGETS 60HR DN | 277 | 166 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS DN | 882 | 538 | All SZGR 2.0 genes in this pathway |
| PURBEY TARGETS OF CTBP1 NOT SATB1 DN | 448 | 282 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION UP | 570 | 339 | All SZGR 2.0 genes in this pathway |