Gene Page: EXOC3
Summary ?
| GeneID | 11336 |
| Symbol | EXOC3 |
| Synonyms | SEC6|SEC6L1|Sec6p |
| Description | exocyst complex component 3 |
| Reference | MIM:608186|HGNC:HGNC:30378|Ensembl:ENSG00000180104|HPRD:09738|Vega:OTTHUMG00000162205 |
| Gene type | protein-coding |
| Map location | 5p15.33 |
| Pascal p-value | 0.634 |
| Sherlock p-value | 0.142 |
| Fetal beta | 0.22 |
| DMG | 1 (# studies) |
| eGene | Anterior cingulate cortex BA24 Caudate basal ganglia Cerebellar Hemisphere Frontal Cortex BA9 Hypothalamus Meta |
| Support | EXOCYTOSIS G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-CONSENSUS G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL G2Cdb.human_BAYES-COLLINS-MOUSE-PSD-CONSENSUS CompositeSet Ascano FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg05479556 | 5 | 449004 | EXOC3 | 4.72E-5 | 0.354 | 0.021 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs72711364 | 5 | 415323 | EXOC3 | ENSG00000180104.11 | 2.108E-6 | 0 | -27950 | gtex_brain_ba24 |
| rs6555242 | 5 | 416546 | EXOC3 | ENSG00000180104.11 | 1.716E-6 | 0 | -26727 | gtex_brain_ba24 |
| rs57433159 | 5 | 418725 | EXOC3 | ENSG00000180104.11 | 1.709E-6 | 0 | -24548 | gtex_brain_ba24 |
| rs72711368 | 5 | 418839 | EXOC3 | ENSG00000180104.11 | 1.578E-6 | 0 | -24434 | gtex_brain_ba24 |
| rs111713756 | 5 | 419318 | EXOC3 | ENSG00000180104.11 | 1.417E-6 | 0 | -23955 | gtex_brain_ba24 |
| rs146034618 | 5 | 420523 | EXOC3 | ENSG00000180104.11 | 1.005E-6 | 0 | -22750 | gtex_brain_ba24 |
| rs72711370 | 5 | 421057 | EXOC3 | ENSG00000180104.11 | 1.096E-6 | 0 | -22216 | gtex_brain_ba24 |
| rs11741946 | 5 | 421403 | EXOC3 | ENSG00000180104.11 | 8.802E-7 | 0 | -21870 | gtex_brain_ba24 |
| rs11741954 | 5 | 421455 | EXOC3 | ENSG00000180104.11 | 1.028E-6 | 0 | -21818 | gtex_brain_ba24 |
| rs10068732 | 5 | 421647 | EXOC3 | ENSG00000180104.11 | 2.008E-9 | 0 | -21626 | gtex_brain_ba24 |
| rs10072668 | 5 | 422752 | EXOC3 | ENSG00000180104.11 | 5.072E-9 | 0 | -20521 | gtex_brain_ba24 |
| rs9313065 | 5 | 423426 | EXOC3 | ENSG00000180104.11 | 5.074E-9 | 0 | -19847 | gtex_brain_ba24 |
| rs13360193 | 5 | 424463 | EXOC3 | ENSG00000180104.11 | 2.601E-9 | 0 | -18810 | gtex_brain_ba24 |
| rs13360218 | 5 | 424612 | EXOC3 | ENSG00000180104.11 | 6.94E-9 | 0 | -18661 | gtex_brain_ba24 |
| rs200261372 | 5 | 426541 | EXOC3 | ENSG00000180104.11 | 3.986E-9 | 0 | -16732 | gtex_brain_ba24 |
| rs12188321 | 5 | 428418 | EXOC3 | ENSG00000180104.11 | 5.072E-9 | 0 | -14855 | gtex_brain_ba24 |
| rs67665078 | 5 | 428754 | EXOC3 | ENSG00000180104.11 | 5.072E-9 | 0 | -14519 | gtex_brain_ba24 |
| rs957791 | 5 | 430217 | EXOC3 | ENSG00000180104.11 | 1.062E-8 | 0 | -13056 | gtex_brain_ba24 |
| rs768479 | 5 | 431196 | EXOC3 | ENSG00000180104.11 | 3.03E-9 | 0 | -12077 | gtex_brain_ba24 |
| rs11740891 | 5 | 435626 | EXOC3 | ENSG00000180104.11 | 7.896E-10 | 0 | -7647 | gtex_brain_ba24 |
| rs5865330 | 5 | 436387 | EXOC3 | ENSG00000180104.11 | 1.837E-9 | 0 | -6886 | gtex_brain_ba24 |
| rs72717425 | 5 | 438809 | EXOC3 | ENSG00000180104.11 | 4.777E-7 | 0 | -4464 | gtex_brain_ba24 |
| rs115554641 | 5 | 443236 | EXOC3 | ENSG00000180104.11 | 6.732E-7 | 0 | -37 | gtex_brain_ba24 |
| rs2721028 | 5 | 443849 | EXOC3 | ENSG00000180104.11 | 1.248E-6 | 0 | 576 | gtex_brain_ba24 |
| rs2037077 | 5 | 447226 | EXOC3 | ENSG00000180104.11 | 1.23E-6 | 0 | 3953 | gtex_brain_ba24 |
| rs2561663 | 5 | 459772 | EXOC3 | ENSG00000180104.11 | 1.985E-6 | 0 | 16499 | gtex_brain_ba24 |
| rs13356700 | 5 | 477719 | EXOC3 | ENSG00000180104.11 | 2.331E-9 | 0 | 34446 | gtex_brain_ba24 |
| rs562360175 | 5 | 478571 | EXOC3 | ENSG00000180104.11 | 7.414E-7 | 0 | 35298 | gtex_brain_ba24 |
| rs9885312 | 5 | 482186 | EXOC3 | ENSG00000180104.11 | 1.601E-6 | 0 | 38913 | gtex_brain_ba24 |
Section II. Transcriptome annotation
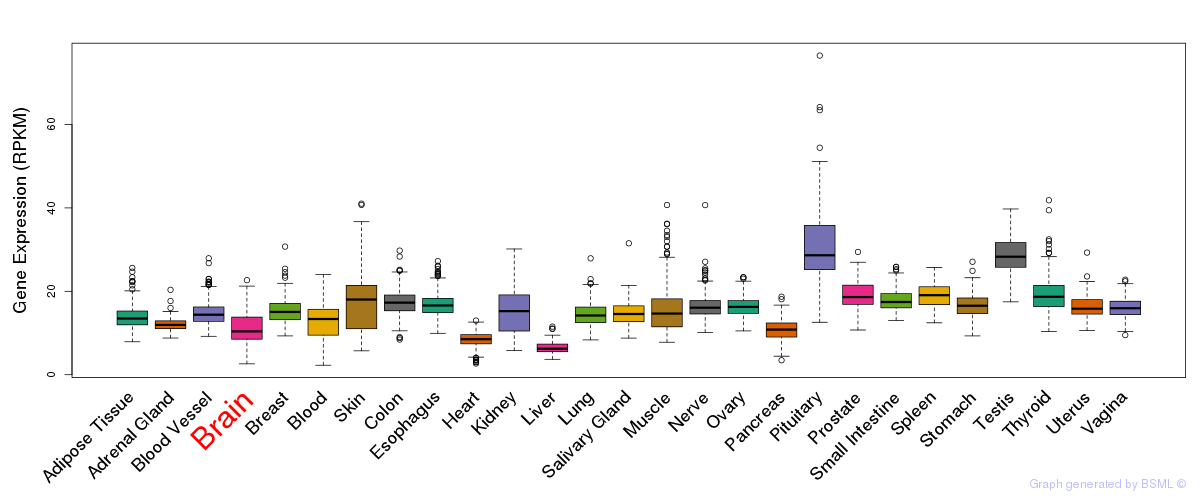
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG TIGHT JUNCTION | 134 | 86 | All SZGR 2.0 genes in this pathway |
| PID INSULIN PATHWAY | 45 | 32 | All SZGR 2.0 genes in this pathway |
| PID ARF6 TRAFFICKING PATHWAY | 49 | 34 | All SZGR 2.0 genes in this pathway |
| PID ECADHERIN STABILIZATION PATHWAY | 42 | 34 | All SZGR 2.0 genes in this pathway |
| REACTOME DIABETES PATHWAYS | 133 | 91 | All SZGR 2.0 genes in this pathway |
| REACTOME INSULIN SYNTHESIS AND PROCESSING | 21 | 15 | All SZGR 2.0 genes in this pathway |
| PARENT MTOR SIGNALING UP | 567 | 375 | All SZGR 2.0 genes in this pathway |
| CASORELLI ACUTE PROMYELOCYTIC LEUKEMIA DN | 663 | 425 | All SZGR 2.0 genes in this pathway |
| WANG LMO4 TARGETS DN | 352 | 225 | All SZGR 2.0 genes in this pathway |
| GINESTIER BREAST CANCER 20Q13 AMPLIFICATION DN | 180 | 101 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY BREAST CANCER 5P15 AMPLICON | 26 | 15 | All SZGR 2.0 genes in this pathway |
| XU GH1 AUTOCRINE TARGETS DN | 142 | 94 | All SZGR 2.0 genes in this pathway |
| AGUIRRE PANCREATIC CANCER COPY NUMBER UP | 298 | 174 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS OLIGODENDROCYTE NUMBER CORR UP | 756 | 494 | All SZGR 2.0 genes in this pathway |
| KIM BIPOLAR DISORDER OLIGODENDROCYTE DENSITY CORR UP | 682 | 440 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS DURATION CORR DN | 146 | 90 | All SZGR 2.0 genes in this pathway |