Gene Page: U2AF2
Summary ?
| GeneID | 11338 |
| Symbol | U2AF2 |
| Synonyms | U2AF65 |
| Description | U2 small nuclear RNA auxiliary factor 2 |
| Reference | MIM:191318|HGNC:HGNC:23156|Ensembl:ENSG00000063244|HPRD:01872|Vega:OTTHUMG00000180908 |
| Gene type | protein-coding |
| Map location | 19q13.42 |
| Pascal p-value | 0.051 |
| Fetal beta | 0.211 |
| DMG | 1 (# studies) |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0583 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg12500884 | 19 | 56165125 | U2AF2 | 7.01E-10 | -0.022 | 9.76E-7 | DMG:Jaffe_2016 |
Section II. Transcriptome annotation
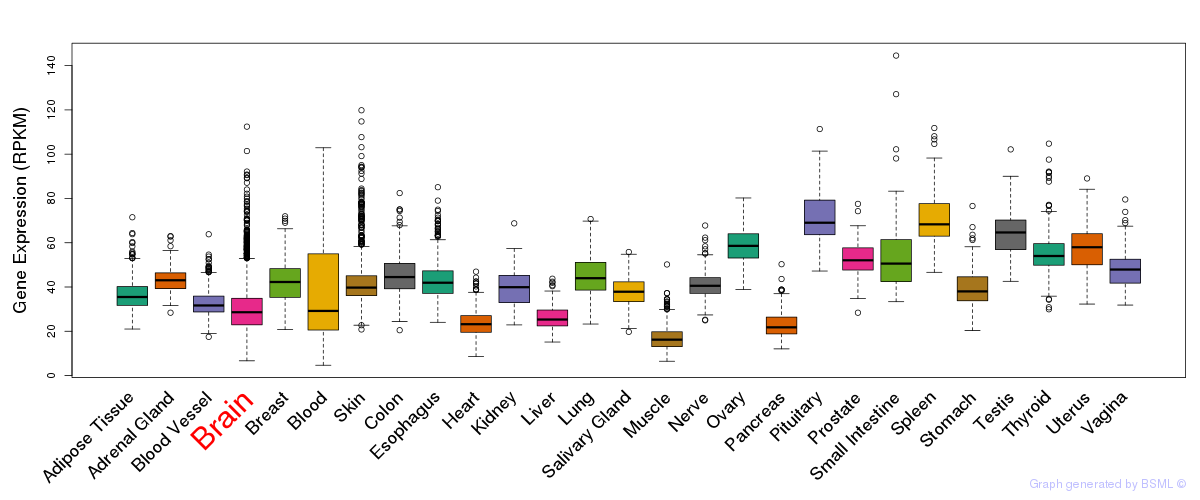
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0003723 | RNA binding | IEA | - | |
| GO:0005515 | protein binding | IPI | 9447963 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0000398 | nuclear mRNA splicing, via spliceosome | EXP | 12226669 | |
| GO:0000398 | nuclear mRNA splicing, via spliceosome | IC | 9731529 | |
| GO:0008380 | RNA splicing | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005634 | nucleus | IEA | - | |
| GO:0005681 | spliceosome | IDA | 9731529 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| BAT1 | D6S81E | DDX39B | UAP56 | HLA-B associated transcript 1 | U2AF65 interacts with UAP56. | BIND | 9242493 |
| BEND7 | C10orf30 | FLJ40283 | MGC35247 | BEN domain containing 7 | Two-hybrid | BioGRID | 16189514 |
| C1orf190 | FLJ25163 | chromosome 1 open reading frame 190 | Two-hybrid | BioGRID | 16189514 |
| CHIC2 | BTL | MGC21173 | cysteine-rich hydrophobic domain 2 | Two-hybrid | BioGRID | 16189514 |
| DDX6 | FLJ36338 | HLR2 | P54 | RCK | DEAD (Asp-Glu-Ala-Asp) box polypeptide 6 | - | HPRD | 8816452 |
| DPPA2 | PESCRG1 | developmental pluripotency associated 2 | Two-hybrid | BioGRID | 16189514 |
| DVL2 | - | dishevelled, dsh homolog 2 (Drosophila) | Two-hybrid | BioGRID | 16189514 |
| KIAA0907 | RP11-336K24.1 | KIAA0907 | Two-hybrid | BioGRID | 16189514 |
| PRPF40A | FBP-11 | FBP11 | FLAF1 | FLJ20585 | FNBP3 | HIP10 | HYPA | NY-REN-6 | PRP40 pre-mRNA processing factor 40 homolog A (S. cerevisiae) | - | HPRD | 15456888 |
| PUF60 | FIR | FLJ31379 | RoBPI | SIAHBP1 | poly-U binding splicing factor 60KDa | Affinity Capture-MS Two-hybrid | BioGRID | 16189514 |17353931 |
| RNPS1 | E5.1 | MGC117332 | RNA binding protein S1, serine-rich domain | Affinity Capture-MS | BioGRID | 17353931 |
| SF1 | D11S636 | ZFM1 | ZNF162 | splicing factor 1 | - | HPRD,BioGRID | 9150140 |9512519 |
| SF1 | D11S636 | ZFM1 | ZNF162 | splicing factor 1 | U2AF65 interacts with an unspecified isoform of SF1 (mBBP). | BIND | 9512519 |
| SF3A2 | PRP11 | PRPF11 | SAP62 | SF3a66 | splicing factor 3a, subunit 2, 66kDa | Affinity Capture-MS | BioGRID | 12234937 |
| SF3B1 | PRP10 | PRPF10 | SAP155 | SF3b155 | splicing factor 3b, subunit 1, 155kDa | SAP155 interacts with U2AF65. | BIND | 9671485 |
| SFRS1 | ASF | MGC5228 | SF2 | SF2p33 | SRp30a | splicing factor, arginine/serine-rich 1 | Two-hybrid | BioGRID | 16189514 |
| SFRS11 | DKFZp686M13204 | dJ677H15.2 | p54 | splicing factor, arginine/serine-rich 11 | - | HPRD,BioGRID | 8816452 |
| SFRS2IP | CASP11 | SIP1 | SRRP129 | splicing factor, arginine/serine-rich 2, interacting protein | - | HPRD,BioGRID | 9447963 |
| SIP1 | GEMIN2 | SIP1-delta | survival of motor neuron protein interacting protein 1 | - | HPRD | 9447963 |
| SRPK1 | SFRSK1 | SFRS protein kinase 1 | - | HPRD | 9472028 |
| SRPK2 | FLJ36101 | SFRSK2 | SFRS protein kinase 2 | Biochemical Activity Two-hybrid | BioGRID | 9472028 |16189514 |
| TCERG1 | CA150 | MGC133200 | TAF2S | transcription elongation regulator 1 | - | HPRD,BioGRID | 15456888 |
| THAP1 | FLJ10477 | MGC33014 | THAP domain containing, apoptosis associated protein 1 | Two-hybrid | BioGRID | 16189514 |
| U2AF1 | DKFZp313J1712 | FP793 | RN | RNU2AF1 | U2AF35 | U2AFBP | U2 small nuclear RNA auxiliary factor 1 | - | HPRD,BioGRID | 1388271 |
| U2AF1 | DKFZp313J1712 | FP793 | RN | RNU2AF1 | U2AF35 | U2AFBP | U2 small nuclear RNA auxiliary factor 1 | hU2AF(65) and hU2AF(35) interact to form the U2AF splice factor heterodimer. | BIND | 9528748 |
| WT1 | GUD | WAGR | WIT-2 | WT33 | Wilms tumor 1 | - | HPRD,BioGRID | 9784496 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG SPLICEOSOME | 128 | 72 | All SZGR 2.0 genes in this pathway |
| REACTOME PROCESSING OF CAPPED INTRON CONTAINING PRE MRNA | 140 | 77 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSPORT OF MATURE TRANSCRIPT TO CYTOPLASM | 54 | 34 | All SZGR 2.0 genes in this pathway |
| REACTOME RNA POL II TRANSCRIPTION | 105 | 54 | All SZGR 2.0 genes in this pathway |
| REACTOME MRNA PROCESSING | 161 | 86 | All SZGR 2.0 genes in this pathway |
| REACTOME MRNA SPLICING | 111 | 58 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSCRIPTION | 210 | 127 | All SZGR 2.0 genes in this pathway |
| REACTOME MRNA 3 END PROCESSING | 35 | 18 | All SZGR 2.0 genes in this pathway |
| REACTOME CLEAVAGE OF GROWING TRANSCRIPT IN THE TERMINATION REGION | 44 | 22 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| GINESTIER BREAST CANCER ZNF217 AMPLIFIED DN | 335 | 193 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA DN | 1375 | 806 | All SZGR 2.0 genes in this pathway |
| XU AKT1 TARGETS 6HR | 27 | 18 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 UP | 309 | 199 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| BENPORATH EED TARGETS | 1062 | 725 | All SZGR 2.0 genes in this pathway |
| SUNG METASTASIS STROMA UP | 110 | 70 | All SZGR 2.0 genes in this pathway |
| MORI EMU MYC LYMPHOMA BY ONSET TIME UP | 110 | 69 | All SZGR 2.0 genes in this pathway |
| TARTE PLASMA CELL VS PLASMABLAST DN | 309 | 206 | All SZGR 2.0 genes in this pathway |
| ASTIER INTEGRIN SIGNALING | 59 | 44 | All SZGR 2.0 genes in this pathway |
| PAL PRMT5 TARGETS UP | 203 | 135 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| JACKSON DNMT1 TARGETS DN | 25 | 21 | All SZGR 2.0 genes in this pathway |
| LEE AGING MUSCLE UP | 45 | 33 | All SZGR 2.0 genes in this pathway |
| AMUNDSON POOR SURVIVAL AFTER GAMMA RADIATION 8G | 95 | 62 | All SZGR 2.0 genes in this pathway |
| BONOME OVARIAN CANCER SURVIVAL SUBOPTIMAL DEBULKING | 510 | 309 | All SZGR 2.0 genes in this pathway |
| GRADE COLON CANCER UP | 871 | 505 | All SZGR 2.0 genes in this pathway |
| AGUIRRE PANCREATIC CANCER COPY NUMBER UP | 298 | 174 | All SZGR 2.0 genes in this pathway |
| HOSHIDA LIVER CANCER LATE RECURRENCE DN | 69 | 48 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH T 8 21 TRANSLOCATION | 368 | 247 | All SZGR 2.0 genes in this pathway |
| KASLER HDAC7 TARGETS 1 UP | 194 | 133 | All SZGR 2.0 genes in this pathway |
| HOLLEMAN PREDNISOLONE RESISTANCE ALL UP | 19 | 15 | All SZGR 2.0 genes in this pathway |