Gene Page: TWF2
Summary ?
| GeneID | 11344 |
| Symbol | TWF2 |
| Synonyms | A6RP|A6r|MSTP011|PTK9L |
| Description | twinfilin actin binding protein 2 |
| Reference | MIM:607433|HGNC:HGNC:9621|Ensembl:ENSG00000247596|HPRD:08464|Vega:OTTHUMG00000158105 |
| Gene type | protein-coding |
| Map location | 3p21.1 |
| Pascal p-value | 1.9E-5 |
| Sherlock p-value | 0.799 |
| Fetal beta | -0.436 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
| LK:YES | Genome-wide Association Study | This data set included 99 genes mapped to the 22 regions. The 24 leading SNPs were also included in CV:Ripke_2013 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg03897318 | 3 | 52272844 | TWF2 | 8.02E-5 | 0.414 | 0.025 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| snp_a-4218218 | 0 | TWF2 | 11344 | 0.13 | trans | |||
| rs584108 | chr11 | 68630281 | TWF2 | 11344 | 0.14 | trans | ||
| rs596874 | chr11 | 68630873 | TWF2 | 11344 | 0.06 | trans | ||
| rs501799 | chr11 | 68631239 | TWF2 | 11344 | 0.18 | trans | ||
| rs646586 | chr11 | 68681652 | TWF2 | 11344 | 0.07 | trans | ||
| rs478647 | chr11 | 68700423 | TWF2 | 11344 | 0.19 | trans | ||
| rs10772528 | chr12 | 12210187 | TWF2 | 11344 | 0.03 | trans |
Section II. Transcriptome annotation
General gene expression (GTEx)
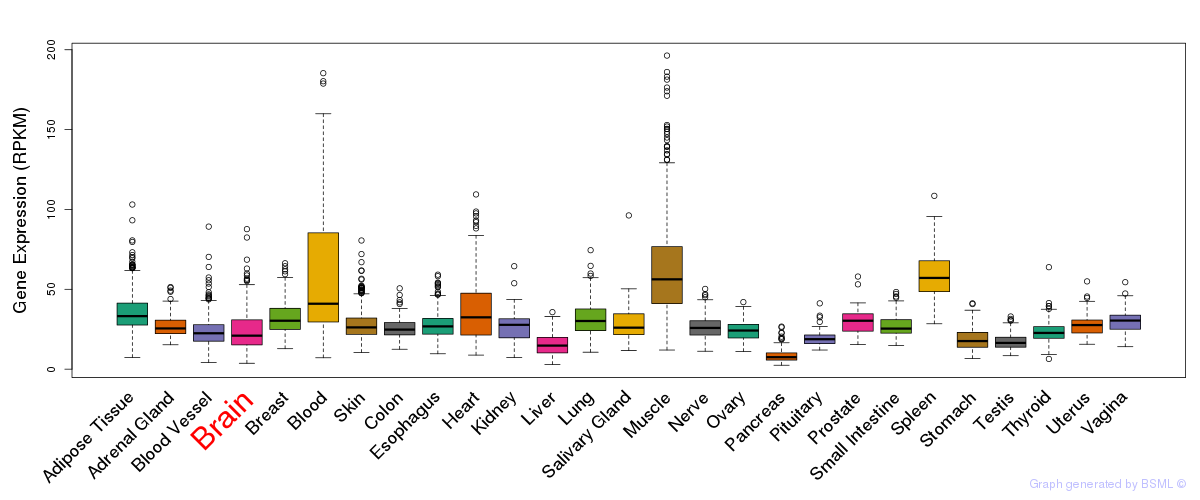
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ACTR3B | ARP11 | ARP3BETA | DKFZp686O24114 | ARP3 actin-related protein 3 homolog B (yeast) | Affinity Capture-MS | BioGRID | 17353931 |
| CAPZA1 | CAPPA1 | CAPZ | CAZ1 | capping protein (actin filament) muscle Z-line, alpha 1 | Affinity Capture-MS | BioGRID | 17353931 |
| CAPZA2 | CAPPA2 | CAPZ | capping protein (actin filament) muscle Z-line, alpha 2 | Affinity Capture-MS | BioGRID | 17353931 |
| CAPZB | CAPB | CAPPB | CAPZ | MGC104401 | MGC129749 | MGC129750 | capping protein (actin filament) muscle Z-line, beta | Affinity Capture-MS | BioGRID | 17353931 |
| CHGB | SCG1 | chromogranin B (secretogranin 1) | Two-hybrid | BioGRID | 16169070 |
| ELOVL1 | CGI-88 | Ssc1 | elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 1 | Affinity Capture-MS | BioGRID | 17353931 |
| ERC1 | Cast2 | ELKS | KIAA1081 | MGC12974 | RAB6IP2 | ELKS/RAB6-interacting/CAST family member 1 | Affinity Capture-MS | BioGRID | 17353931 |
| KRT31 | HA1 | Ha-1 | KRTHA1 | MGC138630 | hHa1 | keratin 31 | Affinity Capture-MS | BioGRID | 17353931 |
| KRT36 | HA6 | KRTHA6 | hHa6 | keratin 36 | Affinity Capture-MS | BioGRID | 17353931 |
| KRT82 | HB2 | Hb-2 | KRTHB2 | keratin 82 | Affinity Capture-MS | BioGRID | 17353931 |
| KRT85 | HB5 | Hb-5 | KRTHB5 | hHb5 | keratin 85 | Affinity Capture-MS | BioGRID | 17353931 |
| LYAR | FLJ20425 | ZLYAR | Ly1 antibody reactive homolog (mouse) | Affinity Capture-MS | BioGRID | 17353931 |
| VRK3 | - | vaccinia related kinase 3 | Affinity Capture-MS | BioGRID | 17353931 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| GAZDA DIAMOND BLACKFAN ANEMIA ERYTHROID DN | 493 | 298 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN UP | 1142 | 669 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND DOXORUBICIN UP | 612 | 367 | All SZGR 2.0 genes in this pathway |
| BORLAK LIVER CANCER EGF UP | 57 | 41 | All SZGR 2.0 genes in this pathway |
| SHETH LIVER CANCER VS TXNIP LOSS PAM1 | 229 | 137 | All SZGR 2.0 genes in this pathway |
| SHETH LIVER CANCER VS TXNIP LOSS PAM2 | 153 | 102 | All SZGR 2.0 genes in this pathway |
| WILLIAMS ESR2 TARGETS UP | 28 | 18 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS DN | 1024 | 594 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 48HR UP | 487 | 286 | All SZGR 2.0 genes in this pathway |
| THEILGAARD NEUTROPHIL AT SKIN WOUND DN | 225 | 163 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS LATE PROGENITOR | 544 | 307 | All SZGR 2.0 genes in this pathway |
| WANG CISPLATIN RESPONSE AND XPC DN | 228 | 146 | All SZGR 2.0 genes in this pathway |
| KAAB HEART ATRIUM VS VENTRICLE DN | 261 | 183 | All SZGR 2.0 genes in this pathway |
| CARDOSO RESPONSE TO GAMMA RADIATION AND 3AB | 18 | 11 | All SZGR 2.0 genes in this pathway |
| STEIN ESRRA TARGETS UP | 388 | 234 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER UP | 973 | 570 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER TUMOR VS NORMAL ADJACENT TISSUE UP | 863 | 514 | All SZGR 2.0 genes in this pathway |
| BONOME OVARIAN CANCER SURVIVAL SUBOPTIMAL DEBULKING | 510 | 309 | All SZGR 2.0 genes in this pathway |
| MOOTHA PGC | 420 | 269 | All SZGR 2.0 genes in this pathway |
| CHEN METABOLIC SYNDROM NETWORK | 1210 | 725 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH 11Q23 REARRANGED | 351 | 238 | All SZGR 2.0 genes in this pathway |
| STEIN ESRRA TARGETS | 535 | 325 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 17 | 181 | 101 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 4HR DN | 254 | 158 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| DELACROIX RAR BOUND ES | 462 | 273 | All SZGR 2.0 genes in this pathway |