Gene Page: XKR4
Summary ?
| GeneID | 114786 |
| Symbol | XKR4 |
| Synonyms | XRG4 |
| Description | XK related 4 |
| Reference | HGNC:HGNC:29394|HPRD:17228| |
| Gene type | protein-coding |
| Map location | 8q12.1 |
| Pascal p-value | 0.123 |
| Sherlock p-value | 0.093 |
| Fetal beta | 0.561 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
General gene expression (GTEx)
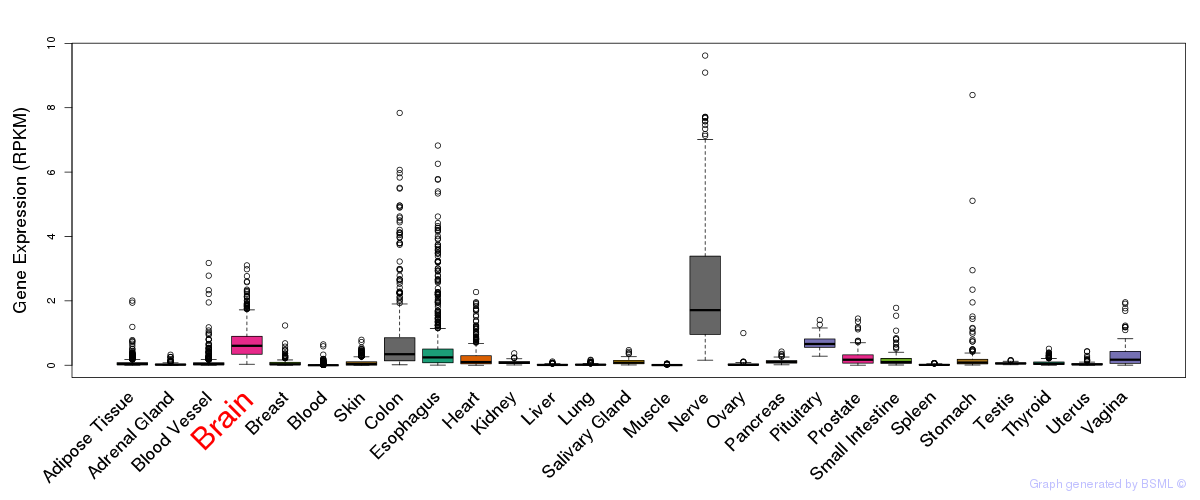
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0016020 | membrane | IEA | - | |
| GO:0016021 | integral to membrane | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| SENESE HDAC3 TARGETS UP | 501 | 327 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION DN | 329 | 219 | All SZGR 2.0 genes in this pathway |
| SABATES COLORECTAL ADENOMA DN | 291 | 176 | All SZGR 2.0 genes in this pathway |
| MEISSNER NPC HCP WITH H3K4ME2 AND H3K27ME3 | 349 | 234 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-129-5p | 804 | 810 | m8 | hsa-miR-129brain | CUUUUUGCGGUCUGGGCUUGC |
| hsa-miR-129-5p | CUUUUUGCGGUCUGGGCUUGCU | ||||
| miR-153 | 712 | 718 | m8 | hsa-miR-153 | UUGCAUAGUCACAAAAGUGA |
| miR-155 | 1420 | 1426 | 1A | hsa-miR-155 | UUAAUGCUAAUCGUGAUAGGGG |
| miR-182 | 1022 | 1028 | 1A | hsa-miR-182 | UUUGGCAAUGGUAGAACUCACA |
| miR-192/215 | 1162 | 1168 | 1A | hsa-miR-192 | CUGACCUAUGAAUUGACAGCC |
| hsa-miR-215 | AUGACCUAUGAAUUGACAGAC | ||||
| miR-208 | 1857 | 1863 | 1A | hsa-miR-208 | AUAAGACGAGCAAAAAGCUUGU |
| miR-29 | 1713 | 1719 | 1A | hsa-miR-29aSZ | UAGCACCAUCUGAAAUCGGUU |
| hsa-miR-29bSZ | UAGCACCAUUUGAAAUCAGUGUU | ||||
| hsa-miR-29cSZ | UAGCACCAUUUGAAAUCGGU | ||||
| hsa-miR-29aSZ | UAGCACCAUCUGAAAUCGGUU | ||||
| hsa-miR-29bSZ | UAGCACCAUUUGAAAUCAGUGUU | ||||
| hsa-miR-29cSZ | UAGCACCAUUUGAAAUCGGU | ||||
| miR-370 | 46 | 52 | m8 | hsa-miR-370brain | GCCUGCUGGGGUGGAACCUGG |
| miR-450 | 804 | 810 | 1A | hsa-miR-450 | UUUUUGCGAUGUGUUCCUAAUA |
| miR-496 | 766 | 772 | 1A | hsa-miR-496 | AUUACAUGGCCAAUCUC |
| miR-499 | 1857 | 1863 | 1A | hsa-miR-499 | UUAAGACUUGCAGUGAUGUUUAA |
| miR-96 | 1021 | 1028 | 1A,m8 | hsa-miR-96brain | UUUGGCACUAGCACAUUUUUGC |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.