Gene Page: GBP4
Summary ?
| GeneID | 115361 |
| Symbol | GBP4 |
| Synonyms | Mpa2 |
| Description | guanylate binding protein 4 |
| Reference | MIM:612466|HGNC:HGNC:20480|Ensembl:ENSG00000162654|HPRD:13570|Vega:OTTHUMG00000010663 |
| Gene type | protein-coding |
| Map location | 1p22.2 |
| Pascal p-value | 0.958 |
| Sherlock p-value | 0.936 |
| DEG p-value | DEG:Sanders_2014:DS1_p=0.160:DS1_beta=0.017100:DS2_p=2.50e-03:DS2_beta=-0.141:DS2_FDR=3.02e-02 |
| Fetal beta | -0.381 |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| DEG:Sanders_2013 | Microarray | Whole-genome gene expression profiles using microarrays on lymphoblastoid cell lines (LCLs) from 413 cases and 446 controls. |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs2745260 | chr1 | 11586676 | GBP4 | 115361 | 0.19 | trans | ||
| rs17109968 | chr1 | 54633542 | GBP4 | 115361 | 0.16 | trans | ||
| rs12036740 | chr1 | 58432174 | GBP4 | 115361 | 0.09 | trans | ||
| rs12041093 | chr1 | 58462509 | GBP4 | 115361 | 0.03 | trans | ||
| rs12023109 | chr1 | 58482942 | GBP4 | 115361 | 0.03 | trans | ||
| rs473019 | chr1 | 72296394 | GBP4 | 115361 | 0.06 | trans | ||
| rs11799385 | chr1 | 79449863 | GBP4 | 115361 | 0.13 | trans | ||
| rs17121311 | chr1 | 100314968 | GBP4 | 115361 | 0.01 | trans | ||
| rs1342707 | chr1 | 147264296 | GBP4 | 115361 | 1.072E-4 | trans | ||
| rs3766034 | chr1 | 169087664 | GBP4 | 115361 | 0 | trans | ||
| rs12748624 | chr1 | 169189770 | GBP4 | 115361 | 0.06 | trans | ||
| rs4027083 | chr1 | 234452908 | GBP4 | 115361 | 0.07 | trans | ||
| rs10495652 | chr2 | 17139288 | GBP4 | 115361 | 1.097E-6 | trans | ||
| rs6531271 | chr2 | 17139932 | GBP4 | 115361 | 1.097E-6 | trans | ||
| rs12620194 | chr2 | 17148003 | GBP4 | 115361 | 1.097E-6 | trans | ||
| rs17042744 | chr2 | 21824821 | GBP4 | 115361 | 0.01 | trans | ||
| rs2113625 | chr2 | 21828617 | GBP4 | 115361 | 0.01 | trans | ||
| rs7563186 | chr2 | 21844897 | GBP4 | 115361 | 1.689E-4 | trans | ||
| rs17680142 | chr2 | 21849670 | GBP4 | 115361 | 0.01 | trans | ||
| rs6547745 | chr2 | 21852551 | GBP4 | 115361 | 0.01 | trans | ||
| rs6758775 | chr2 | 21887142 | GBP4 | 115361 | 0.13 | trans | ||
| rs17042868 | chr2 | 21895688 | GBP4 | 115361 | 0.01 | trans | ||
| rs11893111 | chr2 | 21910980 | GBP4 | 115361 | 0.01 | trans | ||
| rs10490050 | chr2 | 40571949 | GBP4 | 115361 | 0.06 | trans | ||
| snp_a-2186147 | 0 | GBP4 | 115361 | 0.1 | trans | |||
| rs9860881 | chr3 | 40278954 | GBP4 | 115361 | 0.19 | trans | ||
| snp_a-2258988 | 0 | GBP4 | 115361 | 0.01 | trans | |||
| rs17577568 | chr5 | 133167214 | GBP4 | 115361 | 0.06 | trans | ||
| rs10491308 | chr5 | 140678473 | GBP4 | 115361 | 0.03 | trans | ||
| rs10462960 | chr5 | 166125559 | GBP4 | 115361 | 0.01 | trans | ||
| rs2962835 | chr5 | 176331831 | GBP4 | 115361 | 0.01 | trans | ||
| rs10516136 | chr5 | 176381368 | GBP4 | 115361 | 0 | trans | ||
| rs10069302 | chr5 | 176461594 | GBP4 | 115361 | 0.05 | trans | ||
| rs6881260 | chr5 | 176464003 | GBP4 | 115361 | 0.05 | trans | ||
| rs6860827 | chr5 | 176479648 | GBP4 | 115361 | 0.05 | trans | ||
| rs6887893 | chr5 | 176493262 | GBP4 | 115361 | 0.05 | trans | ||
| rs12195984 | 0 | GBP4 | 115361 | 0.15 | trans | |||
| rs16889143 | chr6 | 36757182 | GBP4 | 115361 | 0.03 | trans | ||
| rs16869716 | chr6 | 71743471 | GBP4 | 115361 | 1.585E-6 | trans | ||
| rs1080960 | chr6 | 84815420 | GBP4 | 115361 | 0.15 | trans | ||
| rs13362753 | 0 | GBP4 | 115361 | 0.15 | trans | |||
| rs6455821 | chr6 | 162825101 | GBP4 | 115361 | 0.11 | trans | ||
| rs344468 | chr7 | 146419325 | GBP4 | 115361 | 0.12 | trans | ||
| rs7790094 | chr7 | 152786528 | GBP4 | 115361 | 0.12 | trans | ||
| rs4726262 | chr7 | 152855641 | GBP4 | 115361 | 0.08 | trans | ||
| rs6464313 | chr7 | 152855864 | GBP4 | 115361 | 0.16 | trans | ||
| rs16932340 | chr8 | 66684243 | GBP4 | 115361 | 0 | trans | ||
| rs10827397 | chr10 | 19722202 | GBP4 | 115361 | 0.01 | trans | ||
| rs7907015 | chr10 | 19921035 | GBP4 | 115361 | 0.02 | trans | ||
| rs10827728 | chr10 | 19930366 | GBP4 | 115361 | 0.02 | trans | ||
| rs10509151 | chr10 | 63358651 | GBP4 | 115361 | 0.12 | trans | ||
| rs2393827 | chr10 | 63563590 | GBP4 | 115361 | 7.427E-4 | trans | ||
| rs2588986 | chr10 | 63568258 | GBP4 | 115361 | 7.427E-4 | trans | ||
| rs2053642 | chr10 | 63613118 | GBP4 | 115361 | 0.01 | trans | ||
| rs11027580 | chr11 | 23891659 | GBP4 | 115361 | 0.16 | trans | ||
| rs11027591 | chr11 | 23900616 | GBP4 | 115361 | 0.15 | trans | ||
| rs17133330 | chr11 | 99097728 | GBP4 | 115361 | 0.07 | trans | ||
| rs2592926 | chr11 | 127251272 | GBP4 | 115361 | 0.02 | trans | ||
| rs2618221 | chr11 | 127251893 | GBP4 | 115361 | 0.03 | trans | ||
| rs12577873 | chr11 | 127261987 | GBP4 | 115361 | 0.02 | trans | ||
| rs11220901 | chr11 | 127274538 | GBP4 | 115361 | 0.01 | trans | ||
| rs10893679 | chr11 | 127281378 | GBP4 | 115361 | 0.01 | trans | ||
| rs17395192 | chr12 | 26643930 | GBP4 | 115361 | 0.03 | trans | ||
| rs11613629 | chr12 | 104972976 | GBP4 | 115361 | 0.01 | trans | ||
| rs6492262 | chr13 | 111000751 | GBP4 | 115361 | 0 | trans | ||
| rs7143284 | chr14 | 58320614 | GBP4 | 115361 | 0.02 | trans | ||
| rs927248 | chr14 | 58321206 | GBP4 | 115361 | 0.02 | trans | ||
| rs11158940 | chr14 | 72522541 | GBP4 | 115361 | 0.14 | trans | ||
| rs4774826 | chr15 | 56070006 | GBP4 | 115361 | 6.758E-6 | trans | ||
| rs6498459 | chr16 | 13795898 | GBP4 | 115361 | 0.05 | trans | ||
| rs16995164 | chrX | 148536507 | GBP4 | 115361 | 0.06 | trans |
Section II. Transcriptome annotation
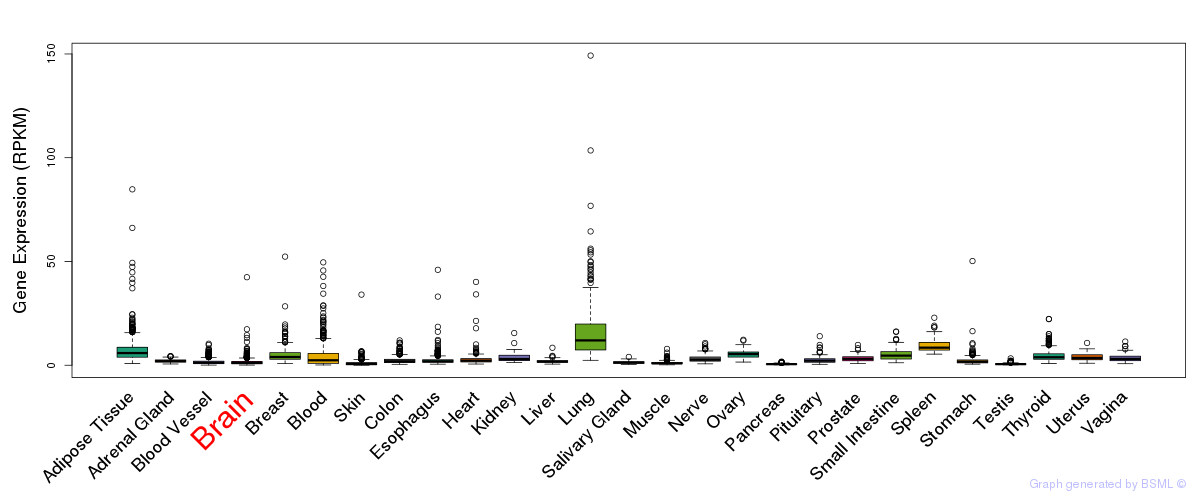
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| REACTOME INTERFERON GAMMA SIGNALING | 63 | 48 | All SZGR 2.0 genes in this pathway |
| REACTOME INTERFERON SIGNALING | 159 | 116 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM | 933 | 616 | All SZGR 2.0 genes in this pathway |
| REACTOME CYTOKINE SIGNALING IN IMMUNE SYSTEM | 270 | 204 | All SZGR 2.0 genes in this pathway |
| BERTUCCI MEDULLARY VS DUCTAL BREAST CANCER UP | 206 | 111 | All SZGR 2.0 genes in this pathway |
| WATANABE ULCERATIVE COLITIS WITH CANCER DN | 14 | 9 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA DN | 1375 | 806 | All SZGR 2.0 genes in this pathway |
| MARKEY RB1 ACUTE LOF UP | 215 | 137 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS UP | 1037 | 673 | All SZGR 2.0 genes in this pathway |
| JECHLINGER EPITHELIAL TO MESENCHYMAL TRANSITION UP | 71 | 51 | All SZGR 2.0 genes in this pathway |
| SANA RESPONSE TO IFNG UP | 78 | 50 | All SZGR 2.0 genes in this pathway |
| YU MYC TARGETS DN | 55 | 38 | All SZGR 2.0 genes in this pathway |
| IGLESIAS E2F TARGETS UP | 151 | 103 | All SZGR 2.0 genes in this pathway |
| HAN JNK SINGALING UP | 35 | 21 | All SZGR 2.0 genes in this pathway |
| LEE AGING CEREBELLUM UP | 84 | 58 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS STEM CELL AND PROGENITOR | 681 | 420 | All SZGR 2.0 genes in this pathway |
| FOSTER TOLERANT MACROPHAGE DN | 409 | 268 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER UP | 973 | 570 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER TUMOR VS NORMAL ADJACENT TISSUE UP | 863 | 514 | All SZGR 2.0 genes in this pathway |
| SEKI INFLAMMATORY RESPONSE LPS UP | 77 | 56 | All SZGR 2.0 genes in this pathway |
| ICHIBA GRAFT VERSUS HOST DISEASE D7 UP | 107 | 67 | All SZGR 2.0 genes in this pathway |
| ICHIBA GRAFT VERSUS HOST DISEASE 35D UP | 131 | 79 | All SZGR 2.0 genes in this pathway |
| SWEET LUNG CANCER KRAS DN | 435 | 289 | All SZGR 2.0 genes in this pathway |
| COLINA TARGETS OF 4EBP1 AND 4EBP2 | 356 | 214 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 15 | 35 | 23 | All SZGR 2.0 genes in this pathway |
| GREGORY SYNTHETIC LETHAL WITH IMATINIB | 145 | 83 | All SZGR 2.0 genes in this pathway |
| CHYLA CBFA2T3 TARGETS UP | 387 | 225 | All SZGR 2.0 genes in this pathway |
| WIERENGA STAT5A TARGETS UP | 217 | 131 | All SZGR 2.0 genes in this pathway |
| WIERENGA STAT5A TARGETS GROUP1 | 136 | 76 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS UP | 745 | 475 | All SZGR 2.0 genes in this pathway |
| SERVITJA ISLET HNF1A TARGETS UP | 163 | 111 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG BOUND 8D | 658 | 397 | All SZGR 2.0 genes in this pathway |
| YANG BCL3 TARGETS UP | 364 | 236 | All SZGR 2.0 genes in this pathway |
| BOSCO INTERFERON INDUCED ANTIVIRAL MODULE | 78 | 48 | All SZGR 2.0 genes in this pathway |
| ALTEMEIER RESPONSE TO LPS WITH MECHANICAL VENTILATION | 128 | 81 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 2ND EGF PULSE ONLY | 1725 | 838 | All SZGR 2.0 genes in this pathway |