Gene Page: CCR3
Summary ?
| GeneID | 1232 |
| Symbol | CCR3 |
| Synonyms | CC-CKR-3|CD193|CKR3|CMKBR3 |
| Description | C-C motif chemokine receptor 3 |
| Reference | MIM:601268|HGNC:HGNC:1604|Ensembl:ENSG00000183625|HPRD:03167|Vega:OTTHUMG00000133484 |
| Gene type | protein-coding |
| Map location | 3p21.3 |
| Pascal p-value | 0.565 |
| Fetal beta | 0.053 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
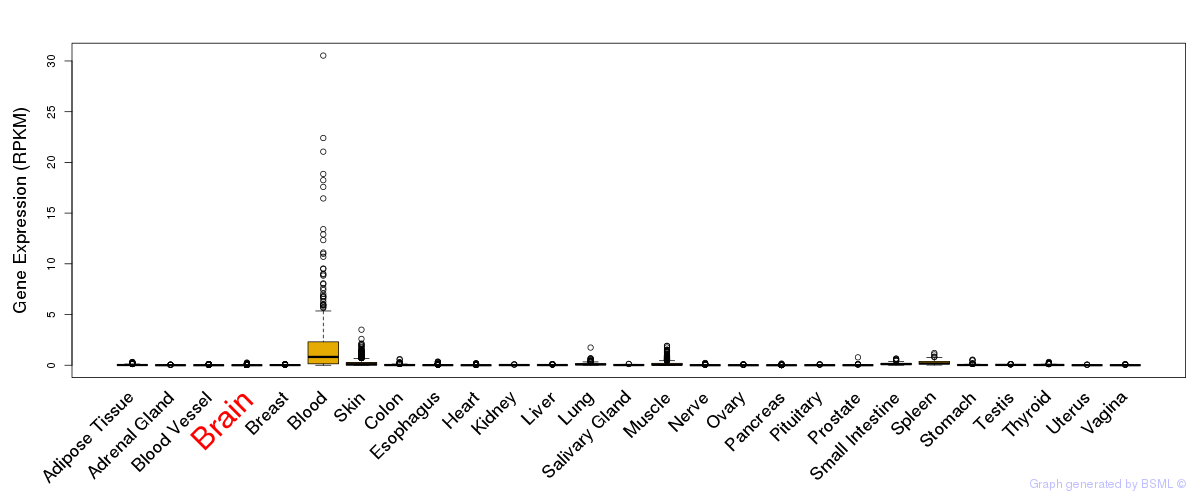
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| LTB4R2 | 0.76 | 0.72 |
| CPNE2 | 0.69 | 0.68 |
| PVRL2 | 0.68 | 0.69 |
| AC100793.2 | 0.66 | 0.58 |
| NPB | 0.66 | 0.59 |
| AC004410.1 | 0.65 | 0.68 |
| MZF1 | 0.64 | 0.64 |
| DUSP9 | 0.64 | 0.53 |
| PAQR7 | 0.63 | 0.64 |
| GPR25 | 0.63 | 0.53 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| KBTBD3 | -0.39 | -0.45 |
| AF347015.27 | -0.37 | -0.37 |
| PLA2G4A | -0.37 | -0.39 |
| AF347015.31 | -0.37 | -0.38 |
| C5orf53 | -0.37 | -0.34 |
| ACOT13 | -0.37 | -0.40 |
| CA4 | -0.36 | -0.32 |
| RERGL | -0.36 | -0.43 |
| MT1G | -0.35 | -0.42 |
| NRN1 | -0.35 | -0.29 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| CCL11 | MGC22554 | SCYA11 | chemokine (C-C motif) ligand 11 | - | HPRD,BioGRID | 8609214 |8642344 |8676064 |9712872 |
| CCL13 | CKb10 | MCP-4 | MGC17134 | NCC-1 | NCC1 | SCYA13 | SCYL1 | chemokine (C-C motif) ligand 13 | - | HPRD,BioGRID | 12149192 |
| CCL14 | CC-1 | CC-3 | CKb1 | HCC-1 | HCC-3 | MCIF | NCC-2 | NCC2 | SCYA14 | SCYL2 | SY14 | chemokine (C-C motif) ligand 14 | - | HPRD,BioGRID | 11085751 |
| CCL15 | HCC-2 | HMRP-2B | LKN1 | Lkn-1 | MIP-1d | MIP-5 | NCC-3 | NCC3 | SCYA15 | SCYL3 | SY15 | chemokine (C-C motif) ligand 15 | - | HPRD | 9346309 |
| CCL2 | GDCF-2 | HC11 | HSMCR30 | MCAF | MCP-1 | MCP1 | MGC9434 | SCYA2 | SMC-CF | chemokine (C-C motif) ligand 2 | Reconstituted Complex | BioGRID | 8642344 |
| CCL24 | Ckb-6 | MPIF-2 | MPIF2 | SCYA24 | chemokine (C-C motif) ligand 24 | - | HPRD | 12218106 |
| CCL26 | IMAC | MGC126714 | MIP-4a | MIP-4alpha | SCYA26 | TSC-1 | chemokine (C-C motif) ligand 26 | - | HPRD | 10488147 |
| CCL28 | CCK1 | MEC | MGC71902 | SCYA28 | chemokine (C-C motif) ligand 28 | - | HPRD | 10975800 |
| CCL3L1 | 464.2 | D17S1718 | G0S19-2 | LD78 | LD78BETA | MGC104178 | MGC12815 | MGC182017 | MIP1AP | SCYA3L | SCYA3L1 | chemokine (C-C motif) ligand 3-like 1 | - | HPRD,BioGRID | 11449371 |
| CCL5 | D17S136E | MGC17164 | RANTES | SCYA5 | SISd | TCP228 | chemokine (C-C motif) ligand 5 | Reconstituted Complex | BioGRID | 8642344 |11449371 |
| CCL5 | D17S136E | MGC17164 | RANTES | SCYA5 | SISd | TCP228 | chemokine (C-C motif) ligand 5 | - | HPRD | 8642344 |9289016 |
| CCL7 | FIC | MARC | MCP-3 | MCP3 | MGC138463 | MGC138465 | NC28 | SCYA6 | SCYA7 | chemokine (C-C motif) ligand 7 | - | HPRD,BioGRID | 8642344 |
| CCL8 | HC14 | MCP-2 | MCP2 | SCYA10 | SCYA8 | chemokine (C-C motif) ligand 8 | - | HPRD | 9005985 |
| FGR | FLJ43153 | MGC75096 | SRC2 | c-fgr | c-src2 | p55c-fgr | p58c-fgr | Gardner-Rasheed feline sarcoma viral (v-fgr) oncogene homolog | - | HPRD,BioGRID | 10527858 |
| HCK | JTK9 | hemopoietic cell kinase | - | HPRD,BioGRID | 10527858 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG CYTOKINE CYTOKINE RECEPTOR INTERACTION | 267 | 161 | All SZGR 2.0 genes in this pathway |
| KEGG CHEMOKINE SIGNALING PATHWAY | 190 | 128 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CCR3 PATHWAY | 23 | 21 | All SZGR 2.0 genes in this pathway |
| BIOCARTA IL5 PATHWAY | 10 | 9 | All SZGR 2.0 genes in this pathway |
| BIOCARTA NKT PATHWAY | 29 | 21 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY GPCR | 920 | 449 | All SZGR 2.0 genes in this pathway |
| REACTOME PEPTIDE LIGAND BINDING RECEPTORS | 188 | 108 | All SZGR 2.0 genes in this pathway |
| REACTOME CLASS A1 RHODOPSIN LIKE RECEPTORS | 305 | 177 | All SZGR 2.0 genes in this pathway |
| REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | 57 | 27 | All SZGR 2.0 genes in this pathway |
| REACTOME GPCR DOWNSTREAM SIGNALING | 805 | 368 | All SZGR 2.0 genes in this pathway |
| REACTOME G ALPHA I SIGNALLING EVENTS | 195 | 114 | All SZGR 2.0 genes in this pathway |
| REACTOME GPCR LIGAND BINDING | 408 | 246 | All SZGR 2.0 genes in this pathway |
| NAKAJIMA EOSINOPHIL | 30 | 20 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 STIMULATED | 1022 | 619 | All SZGR 2.0 genes in this pathway |
| GAURNIER PSMD4 TARGETS | 73 | 55 | All SZGR 2.0 genes in this pathway |
| MAEKAWA ATF2 TARGETS | 24 | 19 | All SZGR 2.0 genes in this pathway |
| ROESSLER LIVER CANCER METASTASIS UP | 107 | 72 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 2ND EGF PULSE ONLY | 1725 | 838 | All SZGR 2.0 genes in this pathway |