Gene Page: CCR5
Summary ?
| GeneID | 1234 |
| Symbol | CCR5 |
| Synonyms | CC-CKR-5|CCCKR5|CCR-5|CD195|CKR-5|CKR5|CMKBR5|IDDM22 |
| Description | C-C motif chemokine receptor 5 (gene/pseudogene) |
| Reference | MIM:601373|HGNC:HGNC:1606|HPRD:03223| |
| Gene type | protein-coding |
| Map location | 3p21.31 |
| Pascal p-value | 0.648 |
| Fetal beta | -0.907 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.4775 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
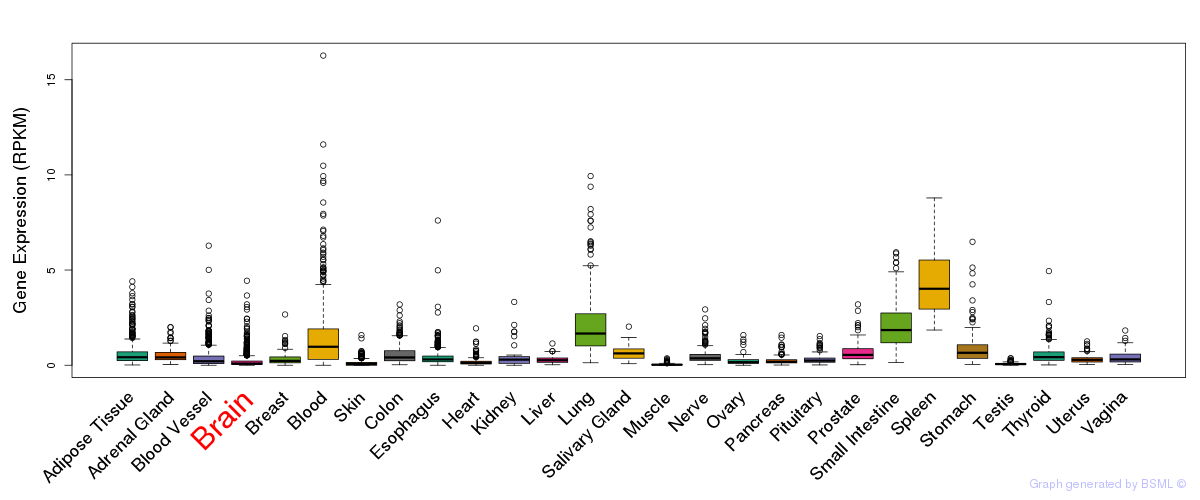
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003779 | actin binding | IDA | 12421915 | |
| GO:0004872 | receptor activity | IEA | - | |
| GO:0005515 | protein binding | IPI | 12421915 |15001559 | |
| GO:0004435 | phosphoinositide phospholipase C activity | TAS | 8663314 | |
| GO:0016493 | C-C chemokine receptor activity | IEA | - | |
| GO:0016493 | C-C chemokine receptor activity | NAS | 8639485 | |
| GO:0015026 | coreceptor activity | TAS | 10469138 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007204 | elevation of cytosolic calcium ion concentration | TAS | 10201901 | |
| GO:0007186 | G-protein coupled receptor protein signaling pathway | IEA | - | |
| GO:0007186 | G-protein coupled receptor protein signaling pathway | TAS | 10393923 | |
| GO:0007267 | cell-cell signaling | TAS | 10679098 | |
| GO:0006968 | cellular defense response | TAS | 10201901 | |
| GO:0007165 | signal transduction | IEA | - | |
| GO:0006955 | immune response | TAS | 8639485 | |
| GO:0006954 | inflammatory response | TAS | 8639485 | |
| GO:0006935 | chemotaxis | TAS | 10741397 | |
| GO:0019059 | initiation of viral infection | EXP | 12091904 | |
| GO:0044419 | interspecies interaction between organisms | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005737 | cytoplasm | TAS | 10415069 | |
| GO:0005768 | endosome | TAS | 10679098 | |
| GO:0016021 | integral to membrane | IEA | - | |
| GO:0009897 | external side of plasma membrane | IDA | 11278962 | |
| GO:0005886 | plasma membrane | IEA | - | |
| GO:0005886 | plasma membrane | TAS | 10201901 | |
| GO:0005887 | integral to plasma membrane | TAS | 10393923 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| AFP | FETA | HPAFP | alpha-fetoprotein | - | HPRD,BioGRID | 12176010 |
| ARRB1 | ARB1 | ARR1 | arrestin, beta 1 | - | HPRD,BioGRID | 12065593 |
| CCL11 | MGC22554 | SCYA11 | chemokine (C-C motif) ligand 11 | - | HPRD | 11264152 |
| CCL13 | CKb10 | MCP-4 | MGC17134 | NCC-1 | NCC1 | SCYA13 | SCYL1 | chemokine (C-C motif) ligand 13 | - | HPRD | 10477718 |
| CCL14 | CC-1 | CC-3 | CKb1 | HCC-1 | HCC-3 | MCIF | NCC-2 | NCC2 | SCYA14 | SCYL2 | SY14 | chemokine (C-C motif) ligand 14 | - | HPRD,BioGRID | 11085751 |
| CCL16 | CKb12 | HCC-4 | ILINCK | LCC-1 | LEC | LMC | MGC117051 | Mtn-1 | NCC-4 | NCC4 | SCYA16 | SCYL4 | chemokine (C-C motif) ligand 16 | - | HPRD,BioGRID | 11470772 |
| CCL2 | GDCF-2 | HC11 | HSMCR30 | MCAF | MCP-1 | MCP1 | MGC9434 | SCYA2 | SMC-CF | chemokine (C-C motif) ligand 2 | - | HPRD | 10477718 |
| CCL3 | G0S19-1 | LD78ALPHA | MIP-1-alpha | MIP1A | SCYA3 | chemokine (C-C motif) ligand 3 | - | HPRD,BioGRID | 11700073 |
| CCL3L1 | 464.2 | D17S1718 | G0S19-2 | LD78 | LD78BETA | MGC104178 | MGC12815 | MGC182017 | MIP1AP | SCYA3L | SCYA3L1 | chemokine (C-C motif) ligand 3-like 1 | - | HPRD,BioGRID | 11449371 |11734558 |
| CCL3L3 | 464.2 | D17S1718 | LD78 | LD78BETA | MGC12815 | SCYA3L | SCYA3L1 | chemokine (C-C motif) ligand 3-like 3 | - | HPRD | 10364178 |11734558 |
| CCL4 | ACT2 | AT744.1 | G-26 | LAG1 | MGC104418 | MGC126025 | MGC126026 | MIP-1-beta | MIP1B | MIP1B1 | SCYA2 | SCYA4 | chemokine (C-C motif) ligand 4 | - | HPRD | 12427015 |
| CCL5 | D17S136E | MGC17164 | RANTES | SCYA5 | SISd | TCP228 | chemokine (C-C motif) ligand 5 | - | HPRD | 8663314 |9289016 |11116158 |14637022 |
| CCL5 | D17S136E | MGC17164 | RANTES | SCYA5 | SISd | TCP228 | chemokine (C-C motif) ligand 5 | Affinity Capture-Western Reconstituted Complex | BioGRID | 11116158 |11449371 |14637022 |
| CCL7 | FIC | MARC | MCP-3 | MCP3 | MGC138463 | MGC138465 | NC28 | SCYA6 | SCYA7 | chemokine (C-C motif) ligand 7 | - | HPRD | 10477718 |
| CCL8 | HC14 | MCP-2 | MCP2 | SCYA10 | SCYA8 | chemokine (C-C motif) ligand 8 | - | HPRD,BioGRID | 9468473 |10477718 |
| CCR2 | CC-CKR-2 | CCR2A | CCR2B | CD192 | CKR2 | CKR2A | CKR2B | CMKBR2 | MCP-1-R | chemokine (C-C motif) receptor 2 | - | HPRD | 11350939 |
| CD4 | CD4mut | CD4 molecule | - | HPRD,BioGRID | 10816381 |
| CXCR4 | CD184 | D2S201E | FB22 | HM89 | HSY3RR | LAP3 | LCR1 | LESTR | NPY3R | NPYR | NPYRL | NPYY3R | WHIM | chemokine (C-X-C motif) receptor 4 | - | HPRD,BioGRID | 12429730 |
| JAK2 | JTK10 | Janus kinase 2 (a protein tyrosine kinase) | - | HPRD | 11591716 |
| JAK2 | JTK10 | Janus kinase 2 (a protein tyrosine kinase) | Affinity Capture-Western | BioGRID | 11278738 |
| LCK | YT16 | p56lck | pp58lck | lymphocyte-specific protein tyrosine kinase | - | HPRD | 11591716 |
| PIK3R2 | P85B | p85 | p85-BETA | phosphoinositide-3-kinase, regulatory subunit 2 (beta) | Affinity Capture-Western | BioGRID | 11350939 |
| PTK2 | FADK | FAK | FAK1 | pp125FAK | PTK2 protein tyrosine kinase 2 | - | HPRD,BioGRID | 10384144 |
| PTK2B | CADTK | CAKB | FADK2 | FAK2 | FRNK | PKB | PTK | PYK2 | RAFTK | PTK2B protein tyrosine kinase 2 beta | - | HPRD | 9446638 |
| STAT3 | APRF | FLJ20882 | HIES | MGC16063 | signal transducer and activator of transcription 3 (acute-phase response factor) | Affinity Capture-Western | BioGRID | 11350939 |
| STAT5B | STAT5 | signal transducer and activator of transcription 5B | Affinity Capture-Western | BioGRID | 11350939 |
| TXK | BTKL | MGC22473 | PSCTK5 | PTK4 | RLK | TKL | TXK tyrosine kinase | - | HPRD | 11859127 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG CYTOKINE CYTOKINE RECEPTOR INTERACTION | 267 | 161 | All SZGR 2.0 genes in this pathway |
| KEGG CHEMOKINE SIGNALING PATHWAY | 190 | 128 | All SZGR 2.0 genes in this pathway |
| KEGG ENDOCYTOSIS | 183 | 132 | All SZGR 2.0 genes in this pathway |
| BIOCARTA TCAPOPTOSIS PATHWAY | 11 | 8 | All SZGR 2.0 genes in this pathway |
| BIOCARTA IL12 PATHWAY | 23 | 17 | All SZGR 2.0 genes in this pathway |
| BIOCARTA NO2IL12 PATHWAY | 17 | 14 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CCR5 PATHWAY | 20 | 16 | All SZGR 2.0 genes in this pathway |
| BIOCARTA NKT PATHWAY | 29 | 21 | All SZGR 2.0 genes in this pathway |
| PID IL12 2PATHWAY | 63 | 54 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY GPCR | 920 | 449 | All SZGR 2.0 genes in this pathway |
| REACTOME PEPTIDE LIGAND BINDING RECEPTORS | 188 | 108 | All SZGR 2.0 genes in this pathway |
| REACTOME CLASS A1 RHODOPSIN LIKE RECEPTORS | 305 | 177 | All SZGR 2.0 genes in this pathway |
| REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | 57 | 27 | All SZGR 2.0 genes in this pathway |
| REACTOME GPCR DOWNSTREAM SIGNALING | 805 | 368 | All SZGR 2.0 genes in this pathway |
| REACTOME G ALPHA I SIGNALLING EVENTS | 195 | 114 | All SZGR 2.0 genes in this pathway |
| REACTOME GPCR LIGAND BINDING | 408 | 246 | All SZGR 2.0 genes in this pathway |
| REACTOME HIV INFECTION | 207 | 122 | All SZGR 2.0 genes in this pathway |
| REACTOME HIV LIFE CYCLE | 125 | 69 | All SZGR 2.0 genes in this pathway |
| REACTOME EARLY PHASE OF HIV LIFE CYCLE | 21 | 8 | All SZGR 2.0 genes in this pathway |
| FULCHER INFLAMMATORY RESPONSE LECTIN VS LPS DN | 463 | 290 | All SZGR 2.0 genes in this pathway |
| ODONNELL TFRC TARGETS UP | 456 | 228 | All SZGR 2.0 genes in this pathway |
| MARKEY RB1 CHRONIC LOF DN | 118 | 78 | All SZGR 2.0 genes in this pathway |
| EBAUER TARGETS OF PAX3 FOXO1 FUSION UP | 207 | 128 | All SZGR 2.0 genes in this pathway |
| FARMER BREAST CANCER CLUSTER 1 | 44 | 23 | All SZGR 2.0 genes in this pathway |
| KANG GIST WITH PDGFRA UP | 50 | 27 | All SZGR 2.0 genes in this pathway |
| SHETH LIVER CANCER VS TXNIP LOSS PAM4 | 261 | 153 | All SZGR 2.0 genes in this pathway |
| LIANG HEMATOPOIESIS STEM CELL NUMBER SMALL VS HUGE UP | 38 | 29 | All SZGR 2.0 genes in this pathway |
| KENNY CTNNB1 TARGETS UP | 50 | 30 | All SZGR 2.0 genes in this pathway |
| MATSUDA NATURAL KILLER DIFFERENTIATION | 475 | 313 | All SZGR 2.0 genes in this pathway |
| ROETH TERT TARGETS DN | 8 | 7 | All SZGR 2.0 genes in this pathway |
| HESS TARGETS OF HOXA9 AND MEIS1 DN | 77 | 48 | All SZGR 2.0 genes in this pathway |
| ZHAN MULTIPLE MYELOMA HP UP | 49 | 26 | All SZGR 2.0 genes in this pathway |
| KAYO CALORIE RESTRICTION MUSCLE UP | 95 | 64 | All SZGR 2.0 genes in this pathway |
| WALLACE PROSTATE CANCER RACE UP | 299 | 167 | All SZGR 2.0 genes in this pathway |
| GAURNIER PSMD4 TARGETS | 73 | 55 | All SZGR 2.0 genes in this pathway |
| CHUNG BLISTER CYTOTOXICITY UP | 134 | 84 | All SZGR 2.0 genes in this pathway |
| BREDEMEYER RAG SIGNALING NOT VIA ATM UP | 62 | 29 | All SZGR 2.0 genes in this pathway |
| QI PLASMACYTOMA UP | 259 | 185 | All SZGR 2.0 genes in this pathway |
| VART KSHV INFECTION ANGIOGENIC MARKERS UP | 165 | 118 | All SZGR 2.0 genes in this pathway |
| GOLDRATH ANTIGEN RESPONSE | 346 | 192 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO CSF2RB AND IL4 DN | 315 | 201 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF UP | 418 | 282 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF VS CSF2RB AND IL4 UP | 408 | 276 | All SZGR 2.0 genes in this pathway |
| SWEET LUNG CANCER KRAS UP | 491 | 316 | All SZGR 2.0 genes in this pathway |
| CHEN METABOLIC SYNDROM NETWORK | 1210 | 725 | All SZGR 2.0 genes in this pathway |
| ONO AML1 TARGETS DN | 41 | 25 | All SZGR 2.0 genes in this pathway |
| ONO FOXP3 TARGETS UP | 23 | 15 | All SZGR 2.0 genes in this pathway |
| KUROZUMI RESPONSE TO ONCOCYTIC VIRUS | 44 | 30 | All SZGR 2.0 genes in this pathway |
| KUROZUMI RESPONSE TO ONCOCYTIC VIRUS AND CYCLIC RGD | 21 | 15 | All SZGR 2.0 genes in this pathway |
| LI INDUCED T TO NATURAL KILLER UP | 307 | 182 | All SZGR 2.0 genes in this pathway |
| VERHAAK GLIOBLASTOMA MESENCHYMAL | 216 | 130 | All SZGR 2.0 genes in this pathway |
| PLASARI TGFB1 SIGNALING VIA NFIC 1HR UP | 33 | 25 | All SZGR 2.0 genes in this pathway |
| BOSCO TH1 CYTOTOXIC MODULE | 114 | 62 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 2ND EGF PULSE ONLY | 1725 | 838 | All SZGR 2.0 genes in this pathway |