Gene Page: COL2A1
Summary ?
| GeneID | 1280 |
| Symbol | COL2A1 |
| Synonyms | ANFH|AOM|COL11A3|SEDC|STL1 |
| Description | collagen type II alpha 1 |
| Reference | MIM:120140|HGNC:HGNC:2200|Ensembl:ENSG00000139219|HPRD:00361|Vega:OTTHUMG00000149896 |
| Gene type | protein-coding |
| Map location | 12q13.11 |
| Pascal p-value | 0.032 |
| Fetal beta | 1 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWAScat | Genome-wide Association Studies | This data set includes 560 SNPs associated with schizophrenia. A total of 486 genes were mapped to these SNPs within 50kb. | |
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg11721803 | 12 | 48398380 | COL2A1 | 2.018E-4 | -0.223 | 0.035 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs10994209 | chr10 | 61877705 | COL2A1 | 1280 | 0.13 | trans |
Section II. Transcriptome annotation
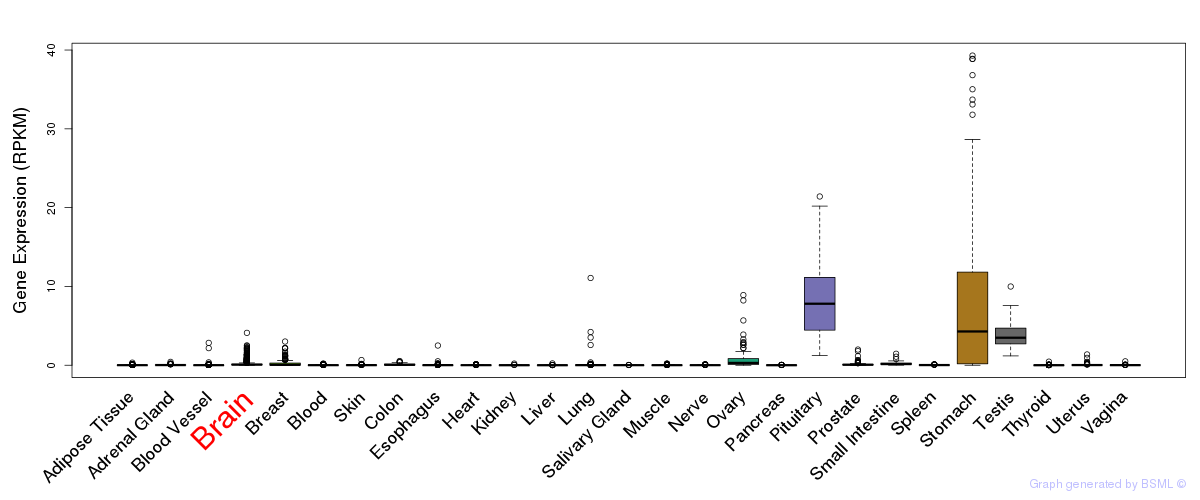
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| NID1 | 0.83 | 0.73 |
| COL11A1 | 0.78 | 0.73 |
| COL1A2 | 0.77 | 0.75 |
| NID2 | 0.76 | 0.73 |
| CPAMD8 | 0.76 | 0.50 |
| ADAMTS12 | 0.76 | 0.60 |
| COL1A1 | 0.76 | 0.62 |
| HSPG2 | 0.76 | 0.70 |
| SVIL | 0.76 | 0.67 |
| EPHB4 | 0.75 | 0.63 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| C5orf53 | -0.37 | -0.43 |
| AF347015.31 | -0.36 | -0.48 |
| AF347015.27 | -0.36 | -0.47 |
| AF347015.33 | -0.36 | -0.46 |
| MT-CO2 | -0.35 | -0.49 |
| S100B | -0.35 | -0.46 |
| MT-CYB | -0.34 | -0.44 |
| FXYD1 | -0.34 | -0.43 |
| AF347015.8 | -0.34 | -0.46 |
| HLA-F | -0.34 | -0.31 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ANXA5 | ANX5 | ENX2 | PP4 | annexin A5 | - | HPRD,BioGRID | 1397263 |
| BGN | DSPG1 | PG-S1 | PGI | SLRR1A | biglycan | - | HPRD,BioGRID | 2059554 |
| BMP2 | BMP2A | bone morphogenetic protein 2 | - | HPRD,BioGRID | 10085302 |
| C1QA | - | complement component 1, q subcomponent, A chain | - | HPRD,BioGRID | 8778019 |
| CHAD | SLRR4A | chondroadherin | - | HPRD | 11445564 |
| COL2A1 | ANFH | AOM | COL11A3 | MGC131516 | SEDC | collagen, type II, alpha 1 | - | HPRD,BioGRID | 1556676 |8660302 |11724554 |
| COL6A1 | OPLL | collagen, type VI, alpha 1 | - | HPRD | 1544908 |
| COL9A1 | DJ149L1.1.2 | EDM6 | FLJ40263 | MED | collagen, type IX, alpha 1 | Reconstituted Complex | BioGRID | 11724554 |
| COL9A2 | DJ39G22.4 | EDM2 | MED | collagen, type IX, alpha 2 | - | HPRD | 11724554 |
| COL9A3 | DJ885L7.4.1 | EDM3 | FLJ90759 | IDD | MED | collagen, type IX, alpha 3 | - | HPRD | 11724554 |
| COMP | EDM1 | EPD1 | MED | MGC131819 | MGC149768 | PSACH | THBS5 | cartilage oligomeric matrix protein | - | HPRD,BioGRID | 9685393 |
| DCN | CSCD | DSPG2 | PG40 | PGII | PGS2 | SLRR1B | decorin | - | HPRD | 10382266 |
| DDR1 | CAK | CD167 | DDR | EDDR1 | MCK10 | NEP | NTRK4 | PTK3 | PTK3A | RTK6 | TRKE | discoidin domain receptor tyrosine kinase 1 | - | HPRD | 9659900 |
| FGF7 | HBGF-7 | KGF | fibroblast growth factor 7 (keratinocyte growth factor) | - | HPRD,BioGRID | 11973338 |
| FN1 | CIG | DKFZp686F10164 | DKFZp686H0342 | DKFZp686I1370 | DKFZp686O13149 | ED-B | FINC | FN | FNZ | GFND | GFND2 | LETS | MSF | fibronectin 1 | - | HPRD,BioGRID | 2229073 |3997552 |
| ITGA2B | CD41 | CD41B | GP2B | GPIIb | GTA | HPA3 | integrin, alpha 2b (platelet glycoprotein IIb of IIb/IIIa complex, antigen CD41) | - | HPRD,BioGRID | 7520045 |10772239 |
| LOXL4 | FLJ21889 | LOXC | lysyl oxidase-like 4 | - | HPRD | 11292829 |
| MAG | GMA | S-MAG | SIGLEC-4A | SIGLEC4A | myelin associated glycoprotein | - | HPRD,BioGRID | 2446864 |
| MATN1 | CMP | CRTM | matrilin 1, cartilage matrix protein | - | HPRD | 10196235 |
| PKD1 | PBP | polycystic kidney disease 1 (autosomal dominant) | - | HPRD,BioGRID | 11406351 |
| PRELP | MGC45323 | MST161 | MSTP161 | SLRR2A | proline/arginine-rich end leucine-rich repeat protein | - | HPRD,BioGRID | 11847210 |
| SPARC | ON | secreted protein, acidic, cysteine-rich (osteonectin) | - | HPRD | 2745554 |
| TGFB1 | CED | DPD1 | TGFB | TGFbeta | transforming growth factor, beta 1 | - | HPRD,BioGRID | 10085302 |
| TGFBI | BIGH3 | CDB1 | CDG2 | CDGG1 | CSD | CSD1 | CSD2 | CSD3 | EBMD | LCD1 | transforming growth factor, beta-induced, 68kDa | - | HPRD | 9061001 |
| THBS1 | THBS | TSP | TSP1 | thrombospondin 1 | - | HPRD,BioGRID | 3571333 |
| TNFRSF10A | APO2 | CD261 | DR4 | MGC9365 | TRAILR-1 | TRAILR1 | tumor necrosis factor receptor superfamily, member 10a | - | HPRD,BioGRID | 9811967 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG FOCAL ADHESION | 201 | 138 | All SZGR 2.0 genes in this pathway |
| KEGG ECM RECEPTOR INTERACTION | 84 | 53 | All SZGR 2.0 genes in this pathway |
| PID INTEGRIN1 PATHWAY | 66 | 44 | All SZGR 2.0 genes in this pathway |
| PID AVB3 INTEGRIN PATHWAY | 75 | 53 | All SZGR 2.0 genes in this pathway |
| PID SYNDECAN 1 PATHWAY | 46 | 29 | All SZGR 2.0 genes in this pathway |
| REACTOME DEVELOPMENTAL BIOLOGY | 396 | 292 | All SZGR 2.0 genes in this pathway |
| REACTOME EXTRACELLULAR MATRIX ORGANIZATION | 87 | 42 | All SZGR 2.0 genes in this pathway |
| REACTOME COLLAGEN FORMATION | 58 | 31 | All SZGR 2.0 genes in this pathway |
| REACTOME INTEGRIN CELL SURFACE INTERACTIONS | 79 | 48 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY PDGF | 122 | 93 | All SZGR 2.0 genes in this pathway |
| REACTOME AXON GUIDANCE | 251 | 188 | All SZGR 2.0 genes in this pathway |
| REACTOME NCAM1 INTERACTIONS | 39 | 27 | All SZGR 2.0 genes in this pathway |
| REACTOME NCAM SIGNALING FOR NEURITE OUT GROWTH | 64 | 49 | All SZGR 2.0 genes in this pathway |
| GAZDA DIAMOND BLACKFAN ANEMIA ERYTHROID DN | 493 | 298 | All SZGR 2.0 genes in this pathway |
| OSWALD HEMATOPOIETIC STEM CELL IN COLLAGEN GEL UP | 233 | 161 | All SZGR 2.0 genes in this pathway |
| ODONNELL TFRC TARGETS UP | 456 | 228 | All SZGR 2.0 genes in this pathway |
| ODONNELL METASTASIS UP | 82 | 58 | All SZGR 2.0 genes in this pathway |
| GAUSSMANN MLL AF4 FUSION TARGETS A DN | 90 | 61 | All SZGR 2.0 genes in this pathway |
| PEREZ TP53 TARGETS | 1174 | 695 | All SZGR 2.0 genes in this pathway |
| HATADA METHYLATED IN LUNG CANCER UP | 390 | 236 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS DN | 848 | 527 | All SZGR 2.0 genes in this pathway |
| BENPORATH SUZ12 TARGETS | 1038 | 678 | All SZGR 2.0 genes in this pathway |
| BENPORATH EED TARGETS | 1062 | 725 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES WITH H3K27ME3 | 1118 | 744 | All SZGR 2.0 genes in this pathway |
| BENPORATH PRC2 TARGETS | 652 | 441 | All SZGR 2.0 genes in this pathway |
| ROSS AML WITH PML RARA FUSION | 77 | 62 | All SZGR 2.0 genes in this pathway |
| POMEROY MEDULLOBLASTOMA DESMOPLASIC VS CLASSIC UP | 62 | 38 | All SZGR 2.0 genes in this pathway |
| LI WILMS TUMOR VS FETAL KIDNEY 1 DN | 163 | 115 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 20HR UP | 240 | 152 | All SZGR 2.0 genes in this pathway |
| KAYO CALORIE RESTRICTION MUSCLE DN | 87 | 59 | All SZGR 2.0 genes in this pathway |
| KAAB HEART ATRIUM VS VENTRICLE UP | 249 | 170 | All SZGR 2.0 genes in this pathway |
| DEBIASI APOPTOSIS BY REOVIRUS INFECTION DN | 287 | 208 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 6HR UP | 71 | 48 | All SZGR 2.0 genes in this pathway |
| KRASNOSELSKAYA ILF3 TARGETS DN | 46 | 38 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 3 | 720 | 440 | All SZGR 2.0 genes in this pathway |
| RIZKI TUMOR INVASIVENESS 3D DN | 270 | 181 | All SZGR 2.0 genes in this pathway |
| FUJII YBX1 TARGETS DN | 202 | 132 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER RELAPSE IN BONE DN | 315 | 197 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER RELAPSE IN LUNG UP | 21 | 10 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL UP | 648 | 398 | All SZGR 2.0 genes in this pathway |
| BOCHKIS FOXA2 TARGETS | 425 | 261 | All SZGR 2.0 genes in this pathway |
| HELLEBREKERS SILENCED DURING TUMOR ANGIOGENESIS | 80 | 56 | All SZGR 2.0 genes in this pathway |
| LI WILMS TUMOR | 27 | 12 | All SZGR 2.0 genes in this pathway |
| SHEDDEN LUNG CANCER POOR SURVIVAL A6 | 456 | 285 | All SZGR 2.0 genes in this pathway |
| COLINA TARGETS OF 4EBP1 AND 4EBP2 | 356 | 214 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K4ME2 AND H3K27ME3 | 59 | 35 | All SZGR 2.0 genes in this pathway |
| MEISSNER NPC HCP WITH H3K4ME2 | 491 | 319 | All SZGR 2.0 genes in this pathway |
| NAKAYAMA FGF2 TARGETS | 29 | 17 | All SZGR 2.0 genes in this pathway |
| HOSHIDA LIVER CANCER SUBCLASS S2 | 115 | 74 | All SZGR 2.0 genes in this pathway |
| NAKAYAMA SOFT TISSUE TUMORS PCA1 DN | 74 | 47 | All SZGR 2.0 genes in this pathway |
| NAKAYAMA SOFT TISSUE TUMORS PCA2 UP | 87 | 50 | All SZGR 2.0 genes in this pathway |
| NIELSEN SYNOVIAL SARCOMA UP | 18 | 14 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 30MIN UP | 56 | 38 | All SZGR 2.0 genes in this pathway |
| WANG METASTASIS OF BREAST CANCER | 15 | 8 | All SZGR 2.0 genes in this pathway |
| KANG AR TARGETS UP | 17 | 10 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS UP | 504 | 321 | All SZGR 2.0 genes in this pathway |
| PURBEY TARGETS OF CTBP1 NOT SATB1 UP | 344 | 215 | All SZGR 2.0 genes in this pathway |
| PLASARI NFIC TARGETS BASAL UP | 27 | 17 | All SZGR 2.0 genes in this pathway |
| PLASARI TGFB1 SIGNALING VIA NFIC 1HR DN | 106 | 77 | All SZGR 2.0 genes in this pathway |
| PLASARI TGFB1 SIGNALING VIA NFIC 10HR UP | 54 | 38 | All SZGR 2.0 genes in this pathway |
| WANG MLL TARGETS | 289 | 188 | All SZGR 2.0 genes in this pathway |
| FOSTER KDM1A TARGETS UP | 266 | 142 | All SZGR 2.0 genes in this pathway |
| NABA COLLAGENS | 44 | 26 | All SZGR 2.0 genes in this pathway |
| NABA CORE MATRISOME | 275 | 148 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME | 1028 | 559 | All SZGR 2.0 genes in this pathway |