Gene Page: COL17A1
Summary ?
| GeneID | 1308 |
| Symbol | COL17A1 |
| Synonyms | BA16H23.2|BP180|BPA-2|BPAG2|ERED|LAD-1 |
| Description | collagen type XVII alpha 1 |
| Reference | MIM:113811|HGNC:HGNC:2194|Ensembl:ENSG00000065618|HPRD:00223|Vega:OTTHUMG00000018998 |
| Gene type | protein-coding |
| Map location | 10q24.3 |
| Pascal p-value | 0.059 |
| eGene | Cerebellum |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
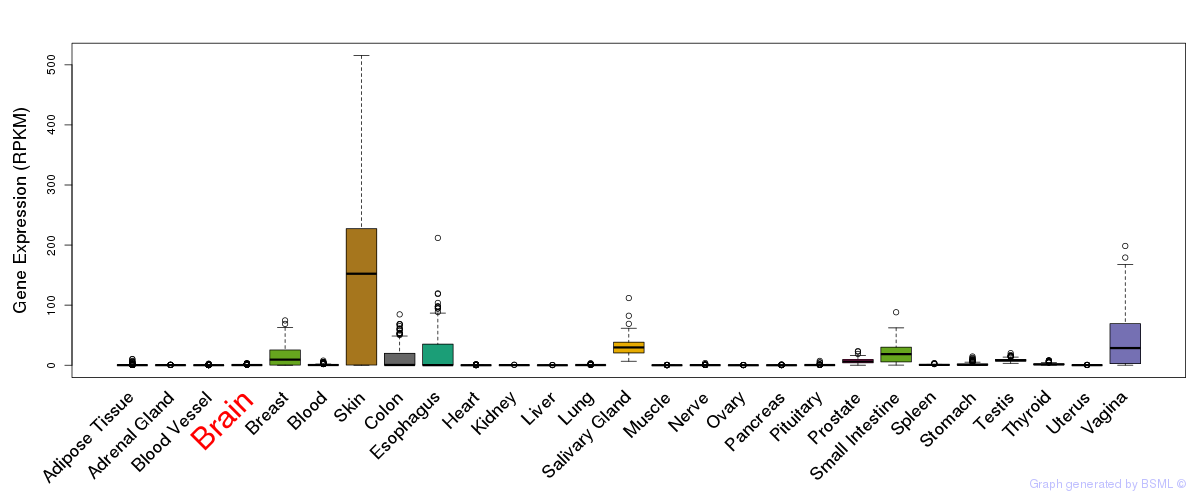
General gene expression (GTEx)
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| HIATL1 | 0.74 | 0.72 |
| DAZAP2 | 0.73 | 0.72 |
| RAB5B | 0.73 | 0.71 |
| STAT5B | 0.73 | 0.73 |
| STAT3 | 0.72 | 0.71 |
| ADAM9 | 0.71 | 0.72 |
| SYT11 | 0.71 | 0.70 |
| TOR1AIP1 | 0.70 | 0.75 |
| SNX1 | 0.70 | 0.70 |
| MPP5 | 0.70 | 0.74 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| RP9P | -0.41 | -0.41 |
| FAM159B | -0.41 | -0.42 |
| AL022328.1 | -0.38 | -0.29 |
| AC135724.1 | -0.37 | -0.35 |
| AF347015.21 | -0.37 | -0.19 |
| AC010300.1 | -0.36 | -0.31 |
| AC087071.1 | -0.35 | -0.29 |
| AC098691.2 | -0.34 | -0.18 |
| AC005921.3 | -0.34 | -0.31 |
| ACSM3 | -0.33 | -0.30 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PID AVB3 INTEGRIN PATHWAY | 75 | 53 | All SZGR 2.0 genes in this pathway |
| PID SYNDECAN 1 PATHWAY | 46 | 29 | All SZGR 2.0 genes in this pathway |
| PID A6B1 A6B4 INTEGRIN PATHWAY | 46 | 35 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CELL COMMUNICATION | 120 | 77 | All SZGR 2.0 genes in this pathway |
| REACTOME EXTRACELLULAR MATRIX ORGANIZATION | 87 | 42 | All SZGR 2.0 genes in this pathway |
| REACTOME COLLAGEN FORMATION | 58 | 31 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL JUNCTION ORGANIZATION | 78 | 43 | All SZGR 2.0 genes in this pathway |
| WATANABE RECTAL CANCER RADIOTHERAPY RESPONSIVE DN | 92 | 51 | All SZGR 2.0 genes in this pathway |
| TURASHVILI BREAST DUCTAL CARCINOMA VS DUCTAL NORMAL DN | 198 | 110 | All SZGR 2.0 genes in this pathway |
| JAEGER METASTASIS DN | 258 | 141 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN UP | 1142 | 669 | All SZGR 2.0 genes in this pathway |
| WATTEL AUTONOMOUS THYROID ADENOMA UP | 73 | 47 | All SZGR 2.0 genes in this pathway |
| ONDER CDH1 TARGETS 2 DN | 464 | 276 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL QTL TRANS | 882 | 572 | All SZGR 2.0 genes in this pathway |
| LEI MYB TARGETS | 318 | 215 | All SZGR 2.0 genes in this pathway |
| CROMER METASTASIS DN | 81 | 58 | All SZGR 2.0 genes in this pathway |
| SESTO RESPONSE TO UV C8 | 72 | 56 | All SZGR 2.0 genes in this pathway |
| LEE AGING CEREBELLUM DN | 86 | 66 | All SZGR 2.0 genes in this pathway |
| JI CARCINOGENESIS BY KRAS AND STK11 UP | 12 | 7 | All SZGR 2.0 genes in this pathway |
| KYNG ENVIRONMENTAL STRESS RESPONSE NOT BY 4NQO IN WS | 40 | 26 | All SZGR 2.0 genes in this pathway |
| KYNG DNA DAMAGE UP | 226 | 164 | All SZGR 2.0 genes in this pathway |
| KYNG ENVIRONMENTAL STRESS RESPONSE DN | 21 | 12 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER NORMAL LIKE UP | 476 | 285 | All SZGR 2.0 genes in this pathway |
| NAKAMURA METASTASIS MODEL UP | 45 | 26 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP B | 549 | 316 | All SZGR 2.0 genes in this pathway |
| KOINUMA TARGETS OF SMAD2 OR SMAD3 | 824 | 528 | All SZGR 2.0 genes in this pathway |
| PEDRIOLI MIR31 TARGETS DN | 418 | 245 | All SZGR 2.0 genes in this pathway |
| ACEVEDO FGFR1 TARGETS IN PROSTATE CANCER MODEL DN | 308 | 187 | All SZGR 2.0 genes in this pathway |
| LIM MAMMARY STEM CELL UP | 489 | 314 | All SZGR 2.0 genes in this pathway |
| NABA COLLAGENS | 44 | 26 | All SZGR 2.0 genes in this pathway |
| NABA CORE MATRISOME | 275 | 148 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME | 1028 | 559 | All SZGR 2.0 genes in this pathway |