Gene Page: COMP
Summary ?
| GeneID | 1311 |
| Symbol | COMP |
| Synonyms | EDM1|EPD1|MED|PSACH|THBS5 |
| Description | cartilage oligomeric matrix protein |
| Reference | MIM:600310|HGNC:HGNC:2227|Ensembl:ENSG00000105664|HPRD:02632|Vega:OTTHUMG00000169318 |
| Gene type | protein-coding |
| Map location | 19p13.1 |
| Pascal p-value | 0.331 |
| DMG | 1 (# studies) |
| eGene | Cerebellum Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenics,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg09980058 | 19 | 18898325 | COMP | 3.38E-4 | -0.406 | 0.041 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs3761082 | chr19 | 19249726 | COMP | 1311 | 0.18 | cis | ||
| rs4653163 | chr1 | 36627444 | COMP | 1311 | 0.04 | trans | ||
| rs12092846 | chr1 | 59940646 | COMP | 1311 | 0 | trans | ||
| rs13447530 | chr1 | 91983798 | COMP | 1311 | 0.01 | trans | ||
| rs6687849 | chr1 | 175904808 | COMP | 1311 | 1.41E-5 | trans | ||
| rs2481653 | chr1 | 176044120 | COMP | 1311 | 0 | trans | ||
| rs2502827 | chr1 | 176044216 | COMP | 1311 | 9.982E-8 | trans | ||
| rs10913839 | chr1 | 179593165 | COMP | 1311 | 0.09 | trans | ||
| rs3008613 | chr1 | 222795768 | COMP | 1311 | 0.15 | trans | ||
| rs882011 | chr2 | 22711668 | COMP | 1311 | 9.828E-4 | trans | ||
| rs17028363 | chr2 | 41913161 | COMP | 1311 | 0 | trans | ||
| rs16829545 | chr2 | 151977407 | COMP | 1311 | 4.03E-16 | trans | ||
| rs16841750 | chr2 | 158288461 | COMP | 1311 | 3.43E-5 | trans | ||
| rs16863816 | chr2 | 177292762 | COMP | 1311 | 0.03 | trans | ||
| rs7584986 | chr2 | 184111432 | COMP | 1311 | 0.03 | trans | ||
| rs10183246 | chr2 | 237187882 | COMP | 1311 | 3.137E-6 | trans | ||
| rs10195546 | chr2 | 237187933 | COMP | 1311 | 1.114E-4 | trans | ||
| rs2322831 | chr3 | 4799093 | COMP | 1311 | 0.03 | trans | ||
| rs4685815 | chr3 | 4799947 | COMP | 1311 | 0.11 | trans | ||
| rs9810143 | chr3 | 5060209 | COMP | 1311 | 3.716E-9 | trans | ||
| rs6797307 | chr3 | 8601563 | COMP | 1311 | 4.634E-6 | trans | ||
| rs6807632 | chr3 | 111395567 | COMP | 1311 | 0 | trans | ||
| rs917937 | chr3 | 137411335 | COMP | 1311 | 0.06 | trans | ||
| rs6773248 | chr3 | 174790333 | COMP | 1311 | 0.01 | trans | ||
| rs6797728 | chr3 | 174790417 | COMP | 1311 | 0.01 | trans | ||
| rs5030115 | chr3 | 186457331 | COMP | 1311 | 0.08 | trans | ||
| rs1996119 | chr3 | 197531387 | COMP | 1311 | 4.925E-4 | trans | ||
| rs16868907 | chr4 | 19933369 | COMP | 1311 | 0.14 | trans | ||
| rs4575988 | chr4 | 19984925 | COMP | 1311 | 0.09 | trans | ||
| rs12642340 | chr4 | 35390674 | COMP | 1311 | 0.13 | trans | ||
| rs662007 | chr4 | 125253160 | COMP | 1311 | 0.02 | trans | ||
| rs1608464 | chr4 | 125318297 | COMP | 1311 | 0.16 | trans | ||
| rs17008119 | chr4 | 125321425 | COMP | 1311 | 6.259E-4 | trans | ||
| rs7692715 | chr4 | 125335209 | COMP | 1311 | 9.863E-5 | trans | ||
| rs6821203 | chr4 | 189475342 | COMP | 1311 | 0.02 | trans | ||
| rs10474151 | chr5 | 84286402 | COMP | 1311 | 0.01 | trans | ||
| rs13360496 | 0 | COMP | 1311 | 0.01 | trans | |||
| rs16890367 | chr6 | 38078448 | COMP | 1311 | 1.077E-5 | trans | ||
| rs12706918 | chr7 | 80266147 | COMP | 1311 | 0.01 | trans | ||
| rs2299269 | chr7 | 95619376 | COMP | 1311 | 0.03 | trans | ||
| rs2166120 | chr7 | 153120003 | COMP | 1311 | 0.1 | trans | ||
| rs6996695 | chr8 | 77540580 | COMP | 1311 | 1.49E-5 | trans | ||
| rs3118341 | chr9 | 25185518 | COMP | 1311 | 0.08 | trans | ||
| rs9406868 | chr9 | 25223372 | COMP | 1311 | 0.1 | trans | ||
| rs17263352 | chr9 | 124811572 | COMP | 1311 | 0.17 | trans | ||
| rs2287296 | chr9 | 132395244 | COMP | 1311 | 0.03 | trans | ||
| rs11005888 | chr10 | 59421572 | COMP | 1311 | 0.14 | trans | ||
| rs17124813 | chr10 | 110347464 | COMP | 1311 | 0.07 | trans | ||
| rs11215990 | chr11 | 116407582 | COMP | 1311 | 0.17 | trans | ||
| rs7972699 | chr12 | 105155290 | COMP | 1311 | 2.714E-6 | trans | ||
| rs7488439 | chr12 | 105155814 | COMP | 1311 | 1.253E-7 | trans | ||
| rs17036489 | chr12 | 105292283 | COMP | 1311 | 1.501E-7 | trans | ||
| rs890460 | chr12 | 119644855 | COMP | 1311 | 0.07 | trans | ||
| rs11060407 | chr12 | 130017919 | COMP | 1311 | 0.17 | trans | ||
| rs11060408 | chr12 | 130018281 | COMP | 1311 | 0.15 | trans | ||
| rs457696 | chr13 | 32581313 | COMP | 1311 | 1.431E-5 | trans | ||
| rs9566986 | chr13 | 32595026 | COMP | 1311 | 7.987E-4 | trans | ||
| rs1530404 | chr13 | 64207519 | COMP | 1311 | 0.07 | trans | ||
| rs17289777 | chr13 | 64223062 | COMP | 1311 | 0.07 | trans | ||
| rs7337531 | chr13 | 109611085 | COMP | 1311 | 0.11 | trans | ||
| rs177369 | chr14 | 73760441 | COMP | 1311 | 0 | trans | ||
| rs1629937 | chr14 | 77059671 | COMP | 1311 | 0.04 | trans | ||
| rs17104720 | chr14 | 77127308 | COMP | 1311 | 0.01 | trans | ||
| rs8009751 | chr14 | 83313138 | COMP | 1311 | 0.06 | trans | ||
| rs16955618 | chr15 | 29937543 | COMP | 1311 | 2.098E-25 | trans | ||
| rs2045264 | chr15 | 94793620 | COMP | 1311 | 0.2 | trans | ||
| rs1420984 | chr16 | 51408733 | COMP | 1311 | 0.01 | trans | ||
| rs16968252 | chr17 | 31692447 | COMP | 1311 | 0.15 | trans | ||
| rs8068272 | chr17 | 49952680 | COMP | 1311 | 0.17 | trans | ||
| rs17820517 | chr17 | 53980462 | COMP | 1311 | 0.01 | trans | ||
| rs17758736 | chr17 | 54035819 | COMP | 1311 | 0.02 | trans | ||
| rs17823633 | chr17 | 68851458 | COMP | 1311 | 2.524E-4 | trans | ||
| rs755768 | chr17 | 75023387 | COMP | 1311 | 0.02 | trans | ||
| rs11873184 | chr18 | 1584081 | COMP | 1311 | 0.06 | trans | ||
| rs17681878 | chr18 | 34767223 | COMP | 1311 | 0.14 | trans | ||
| rs17071138 | chr18 | 61143907 | COMP | 1311 | 0.1 | trans | ||
| rs6032295 | chr20 | 44178138 | COMP | 1311 | 0.02 | trans | ||
| rs6019060 | chr20 | 46965832 | COMP | 1311 | 0.01 | trans | ||
| rs749043 | chr20 | 46995700 | COMP | 1311 | 9.253E-6 | trans | ||
| rs1041786 | chr21 | 22617710 | COMP | 1311 | 0.01 | trans | ||
| rs7278206 | chr21 | 28171971 | COMP | 1311 | 0.18 | trans | ||
| rs5992807 | chr22 | 18210821 | COMP | 1311 | 0.07 | trans | ||
| rs7288523 | chr22 | 50082517 | COMP | 1311 | 0.15 | trans | ||
| rs16990575 | chrX | 32691327 | COMP | 1311 | 3.93E-4 | trans | ||
| rs16993189 | chrX | 144666166 | COMP | 1311 | 0 | trans |
Section II. Transcriptome annotation
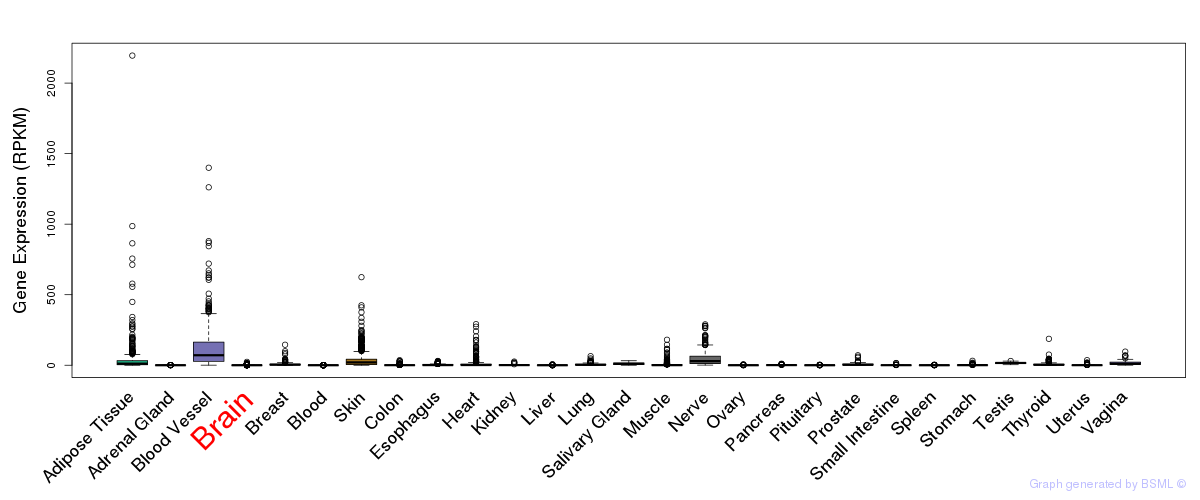
General gene expression (GTEx)
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG TGF BETA SIGNALING PATHWAY | 86 | 64 | All SZGR 2.0 genes in this pathway |
| KEGG FOCAL ADHESION | 201 | 138 | All SZGR 2.0 genes in this pathway |
| KEGG ECM RECEPTOR INTERACTION | 84 | 53 | All SZGR 2.0 genes in this pathway |
| SCHUETZ BREAST CANCER DUCTAL INVASIVE UP | 351 | 230 | All SZGR 2.0 genes in this pathway |
| TURASHVILI BREAST LOBULAR CARCINOMA VS DUCTAL NORMAL UP | 69 | 38 | All SZGR 2.0 genes in this pathway |
| TURASHVILI BREAST LOBULAR CARCINOMA VS LOBULAR NORMAL DN | 74 | 42 | All SZGR 2.0 genes in this pathway |
| DELYS THYROID CANCER UP | 443 | 294 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN UP | 1142 | 669 | All SZGR 2.0 genes in this pathway |
| KIM MYCN AMPLIFICATION TARGETS UP | 92 | 64 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| ROPERO HDAC2 TARGETS | 114 | 71 | All SZGR 2.0 genes in this pathway |
| RICKMAN HEAD AND NECK CANCER B | 48 | 22 | All SZGR 2.0 genes in this pathway |
| BENPORATH SUZ12 TARGETS | 1038 | 678 | All SZGR 2.0 genes in this pathway |
| BENPORATH EED TARGETS | 1062 | 725 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES WITH H3K27ME3 | 1118 | 744 | All SZGR 2.0 genes in this pathway |
| BENPORATH PRC2 TARGETS | 652 | 441 | All SZGR 2.0 genes in this pathway |
| ASTON MAJOR DEPRESSIVE DISORDER UP | 49 | 36 | All SZGR 2.0 genes in this pathway |
| TRAYNOR RETT SYNDROM DN | 19 | 14 | All SZGR 2.0 genes in this pathway |
| KAAB HEART ATRIUM VS VENTRICLE UP | 249 | 170 | All SZGR 2.0 genes in this pathway |
| RODWELL AGING KIDNEY UP | 487 | 303 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 UP | 428 | 266 | All SZGR 2.0 genes in this pathway |
| LY AGING MIDDLE UP | 14 | 11 | All SZGR 2.0 genes in this pathway |
| MITSIADES RESPONSE TO APLIDIN UP | 439 | 257 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER RELAPSE IN LUNG DN | 37 | 22 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL DN | 701 | 446 | All SZGR 2.0 genes in this pathway |
| BOQUEST STEM CELL CULTURED VS FRESH UP | 425 | 298 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K4ME2 AND H3K27ME3 | 59 | 35 | All SZGR 2.0 genes in this pathway |
| MEISSNER NPC HCP WITH H3K4ME2 | 491 | 319 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MCV6 HCP WITH H3K27ME3 | 435 | 318 | All SZGR 2.0 genes in this pathway |
| LY AGING OLD UP | 7 | 5 | All SZGR 2.0 genes in this pathway |
| PANGAS TUMOR SUPPRESSION BY SMAD1 AND SMAD5 DN | 157 | 106 | All SZGR 2.0 genes in this pathway |
| ANASTASSIOU CANCER MESENCHYMAL TRANSITION SIGNATURE | 64 | 40 | All SZGR 2.0 genes in this pathway |
| WARTERS RESPONSE TO IR SKIN | 83 | 44 | All SZGR 2.0 genes in this pathway |
| NABA ECM GLYCOPROTEINS | 196 | 99 | All SZGR 2.0 genes in this pathway |
| NABA CORE MATRISOME | 275 | 148 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME | 1028 | 559 | All SZGR 2.0 genes in this pathway |