Gene Page: MAP3K8
Summary ?
| GeneID | 1326 |
| Symbol | MAP3K8 |
| Synonyms | AURA2|COT|EST|ESTF|MEKK8|TPL2|Tpl-2|c-COT |
| Description | mitogen-activated protein kinase kinase kinase 8 |
| Reference | MIM:191195|HGNC:HGNC:6860|Ensembl:ENSG00000107968|HPRD:01863|Vega:OTTHUMG00000017889 |
| Gene type | protein-coding |
| Map location | 10p11.23 |
| Pascal p-value | 0.88 |
| Fetal beta | 0.216 |
| DMG | 1 (# studies) |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0333 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg04081651 | 10 | 30722660 | MAP3K8 | 3.2E-10 | -0.016 | 7.03E-7 | DMG:Jaffe_2016 |
Section II. Transcriptome annotation
General gene expression (GTEx)
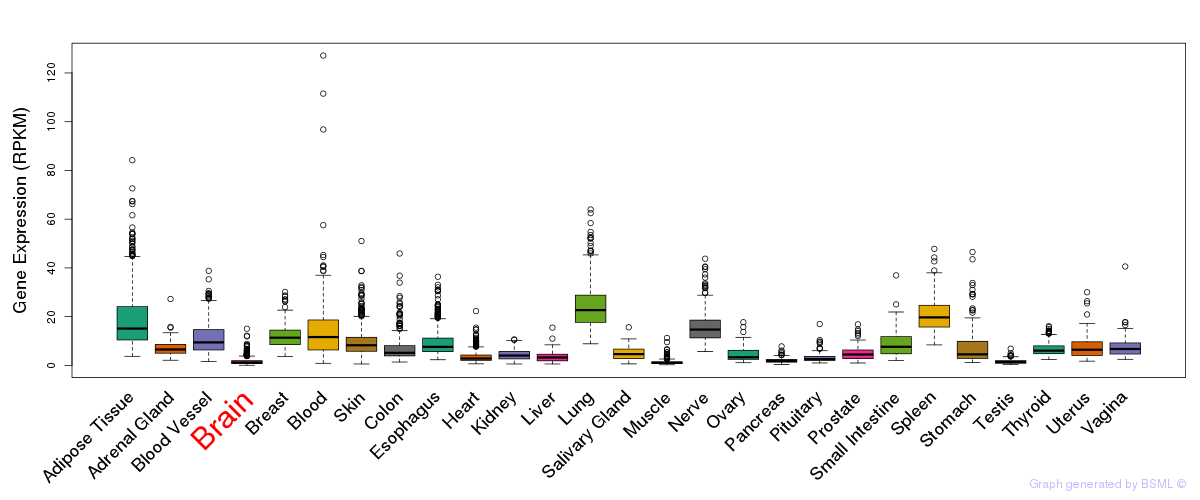
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0000287 | magnesium ion binding | IEA | - | |
| GO:0005515 | protein binding | IEA | - | |
| GO:0005515 | protein binding | IPI | 15169888 | |
| GO:0005524 | ATP binding | IEA | - | |
| GO:0004672 | protein kinase activity | IEA | - | |
| GO:0004674 | protein serine/threonine kinase activity | TAS | 8226782 | |
| GO:0004709 | MAP kinase kinase kinase activity | IEA | - | |
| GO:0016740 | transferase activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006468 | protein amino acid phosphorylation | IEA | - | |
| GO:0006468 | protein amino acid phosphorylation | TAS | 2072910 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005829 | cytosol | TAS | 8226782 | |
| GO:0005737 | cytoplasm | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| AKT1 | AKT | MGC99656 | PKB | PKB-ALPHA | PRKBA | RAC | RAC-ALPHA | v-akt murine thymoma viral oncogene homolog 1 | - | HPRD,BioGRID | 12138205 |
| CHUK | IKBKA | IKK-alpha | IKK1 | IKKA | NFKBIKA | TCF16 | conserved helix-loop-helix ubiquitous kinase | - | HPRD,BioGRID | 10072079 |
| KSR2 | FLJ25965 | kinase suppressor of ras 2 | Affinity Capture-Western | BioGRID | 12975377 |
| MAP2K1 | MAPKK1 | MEK1 | MKK1 | PRKMK1 | mitogen-activated protein kinase kinase 1 | Biochemical Activity | BioGRID | 8631303 |
| MAP2K4 | JNKK | JNKK1 | MAPKK4 | MEK4 | MKK4 | PRKMK4 | SEK1 | SERK1 | mitogen-activated protein kinase kinase 4 | - | HPRD,BioGRID | 8631303 |
| MAP3K14 | FTDCR1B | HS | HSNIK | NIK | mitogen-activated protein kinase kinase kinase 14 | - | HPRD,BioGRID | 10072079 |
| NFKB1 | DKFZp686C01211 | EBP-1 | KBF1 | MGC54151 | NF-kappa-B | NFKB-p105 | NFKB-p50 | p105 | nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 | - | HPRD,BioGRID | 9950430 |
| NFKB2 | LYT-10 | LYT10 | nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100) | Affinity Capture-MS | BioGRID | 14743216 |
| REL | C-Rel | v-rel reticuloendotheliosis viral oncogene homolog (avian) | Affinity Capture-MS | BioGRID | 14743216 |
| RELA | MGC131774 | NFKB3 | p65 | v-rel reticuloendotheliosis viral oncogene homolog A (avian) | Affinity Capture-MS | BioGRID | 14743216 |
| TNIP2 | ABIN-2 | ABIN2 | FLIP1 | KLIP | MGC4289 | TNFAIP3 interacting protein 2 | - | HPRD,BioGRID | 14743216 |15169888 |
| TRAF2 | MGC:45012 | TRAP | TRAP3 | TNF receptor-associated factor 2 | - | HPRD,BioGRID | 11932422 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG MAPK SIGNALING PATHWAY | 267 | 205 | All SZGR 2.0 genes in this pathway |
| KEGG TOLL LIKE RECEPTOR SIGNALING PATHWAY | 102 | 88 | All SZGR 2.0 genes in this pathway |
| KEGG T CELL RECEPTOR SIGNALING PATHWAY | 108 | 89 | All SZGR 2.0 genes in this pathway |
| BIOCARTA MAPK PATHWAY | 87 | 68 | All SZGR 2.0 genes in this pathway |
| ST ERK1 ERK2 MAPK PATHWAY | 32 | 25 | All SZGR 2.0 genes in this pathway |
| PID TCR PATHWAY | 66 | 51 | All SZGR 2.0 genes in this pathway |
| PID CD8 TCR PATHWAY | 53 | 42 | All SZGR 2.0 genes in this pathway |
| PID NFAT 3PATHWAY | 54 | 47 | All SZGR 2.0 genes in this pathway |
| PID TCR RAS PATHWAY | 14 | 14 | All SZGR 2.0 genes in this pathway |
| PID TCR JNK PATHWAY | 14 | 11 | All SZGR 2.0 genes in this pathway |
| REACTOME CD28 CO STIMULATION | 32 | 26 | All SZGR 2.0 genes in this pathway |
| REACTOME COSTIMULATION BY THE CD28 FAMILY | 63 | 48 | All SZGR 2.0 genes in this pathway |
| REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | 22 | 18 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY ILS | 107 | 86 | All SZGR 2.0 genes in this pathway |
| REACTOME IL1 SIGNALING | 39 | 30 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM | 933 | 616 | All SZGR 2.0 genes in this pathway |
| REACTOME ADAPTIVE IMMUNE SYSTEM | 539 | 350 | All SZGR 2.0 genes in this pathway |
| REACTOME CYTOKINE SIGNALING IN IMMUNE SYSTEM | 270 | 204 | All SZGR 2.0 genes in this pathway |
| PICCALUGA ANGIOIMMUNOBLASTIC LYMPHOMA DN | 136 | 86 | All SZGR 2.0 genes in this pathway |
| WILCOX RESPONSE TO PROGESTERONE UP | 152 | 90 | All SZGR 2.0 genes in this pathway |
| FULCHER INFLAMMATORY RESPONSE LECTIN VS LPS DN | 463 | 290 | All SZGR 2.0 genes in this pathway |
| RHEIN ALL GLUCOCORTICOID THERAPY UP | 78 | 41 | All SZGR 2.0 genes in this pathway |
| ODONNELL TFRC TARGETS UP | 456 | 228 | All SZGR 2.0 genes in this pathway |
| ODONNELL TARGETS OF MYC AND TFRC UP | 83 | 50 | All SZGR 2.0 genes in this pathway |
| NAGASHIMA NRG1 SIGNALING UP | 176 | 123 | All SZGR 2.0 genes in this pathway |
| WAMUNYOKOLI OVARIAN CANCER LMP DN | 199 | 124 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE EPIDERMIS DN | 508 | 354 | All SZGR 2.0 genes in this pathway |
| GAUSSMANN MLL AF4 FUSION TARGETS F UP | 185 | 119 | All SZGR 2.0 genes in this pathway |
| RASHI RESPONSE TO IONIZING RADIATION 2 | 127 | 92 | All SZGR 2.0 genes in this pathway |
| RASHI NFKB1 TARGETS | 19 | 18 | All SZGR 2.0 genes in this pathway |
| SCHEIDEREIT IKK INTERACTING PROTEINS | 58 | 45 | All SZGR 2.0 genes in this pathway |
| SHI SPARC TARGETS DN | 13 | 8 | All SZGR 2.0 genes in this pathway |
| THEODOROU MAMMARY TUMORIGENESIS | 31 | 24 | All SZGR 2.0 genes in this pathway |
| AMIT SERUM RESPONSE 60 MCF10A | 57 | 42 | All SZGR 2.0 genes in this pathway |
| SUNG METASTASIS STROMA UP | 110 | 70 | All SZGR 2.0 genes in this pathway |
| MORI MATURE B LYMPHOCYTE UP | 90 | 62 | All SZGR 2.0 genes in this pathway |
| BROCKE APOPTOSIS REVERSED BY IL6 | 144 | 98 | All SZGR 2.0 genes in this pathway |
| SWEET KRAS TARGETS UP | 84 | 51 | All SZGR 2.0 genes in this pathway |
| BASSO CD40 SIGNALING UP | 101 | 76 | All SZGR 2.0 genes in this pathway |
| YIH RESPONSE TO ARSENITE C4 | 18 | 12 | All SZGR 2.0 genes in this pathway |
| RODWELL AGING KIDNEY UP | 487 | 303 | All SZGR 2.0 genes in this pathway |
| WANG SMARCE1 TARGETS UP | 280 | 183 | All SZGR 2.0 genes in this pathway |
| GAVIN FOXP3 TARGETS CLUSTER P4 | 100 | 62 | All SZGR 2.0 genes in this pathway |
| GAVIN IL2 RESPONSIVE FOXP3 TARGETS UP | 19 | 12 | All SZGR 2.0 genes in this pathway |
| ZHENG BOUND BY FOXP3 | 491 | 310 | All SZGR 2.0 genes in this pathway |
| ZHENG FOXP3 TARGETS UP | 26 | 16 | All SZGR 2.0 genes in this pathway |
| FOSTER TOLERANT MACROPHAGE DN | 409 | 268 | All SZGR 2.0 genes in this pathway |
| KONDO EZH2 TARGETS | 245 | 148 | All SZGR 2.0 genes in this pathway |
| HELLER SILENCED BY METHYLATION UP | 282 | 183 | All SZGR 2.0 genes in this pathway |
| ENGELMANN CANCER PROGENITORS UP | 48 | 31 | All SZGR 2.0 genes in this pathway |
| GRADE METASTASIS DN | 45 | 31 | All SZGR 2.0 genes in this pathway |
| BOQUEST STEM CELL CULTURED VS FRESH UP | 425 | 298 | All SZGR 2.0 genes in this pathway |
| BREDEMEYER RAG SIGNALING VIA ATM NOT VIA NFKB UP | 49 | 32 | All SZGR 2.0 genes in this pathway |
| FIRESTEIN PROLIFERATION | 175 | 125 | All SZGR 2.0 genes in this pathway |
| CHANG CORE SERUM RESPONSE UP | 212 | 128 | All SZGR 2.0 genes in this pathway |
| CHEN METABOLIC SYNDROM NETWORK | 1210 | 725 | All SZGR 2.0 genes in this pathway |
| CHIANG LIVER CANCER SUBCLASS CTNNB1 UP | 176 | 110 | All SZGR 2.0 genes in this pathway |
| UZONYI RESPONSE TO LEUKOTRIENE AND THROMBIN | 37 | 26 | All SZGR 2.0 genes in this pathway |
| CHANDRAN METASTASIS UP | 221 | 135 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 1 | 68 | 50 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 2HR UP | 39 | 30 | All SZGR 2.0 genes in this pathway |
| DUTERTRE ESTRADIOL RESPONSE 24HR DN | 505 | 328 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP A | 898 | 516 | All SZGR 2.0 genes in this pathway |
| ALTEMEIER RESPONSE TO LPS WITH MECHANICAL VENTILATION | 128 | 81 | All SZGR 2.0 genes in this pathway |
| ZWANG CLASS 3 TRANSIENTLY INDUCED BY EGF | 222 | 159 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-140 | 680 | 687 | 1A,m8 | hsa-miR-140brain | AGUGGUUUUACCCUAUGGUAG |
| miR-146 | 199 | 205 | 1A | hsa-miR-146a | UGAGAACUGAAUUCCAUGGGUU |
| hsa-miR-146bbrain | UGAGAACUGAAUUCCAUAGGCU | ||||
| miR-17-5p/20/93.mr/106/519.d | 286 | 292 | m8 | hsa-miR-17-5p | CAAAGUGCUUACAGUGCAGGUAGU |
| hsa-miR-20abrain | UAAAGUGCUUAUAGUGCAGGUAG | ||||
| hsa-miR-106a | AAAAGUGCUUACAGUGCAGGUAGC | ||||
| hsa-miR-106bSZ | UAAAGUGCUGACAGUGCAGAU | ||||
| hsa-miR-20bSZ | CAAAGUGCUCAUAGUGCAGGUAG | ||||
| hsa-miR-519d | CAAAGUGCCUCCCUUUAGAGUGU | ||||
| miR-299-3p | 683 | 689 | 1A | hsa-miR-299-3p | UAUGUGGGAUGGUAAACCGCUU |
| miR-299-5p | 678 | 685 | 1A,m8 | hsa-miR-299-5p | UGGUUUACCGUCCCACAUACAU |
| miR-370 | 567 | 573 | m8 | hsa-miR-370brain | GCCUGCUGGGGUGGAACCUGG |
| miR-539 | 205 | 212 | 1A,m8 | hsa-miR-539 | GGAGAAAUUAUCCUUGGUGUGU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.