Gene Page: ADORA2A
Summary ?
| GeneID | 135 |
| Symbol | ADORA2A |
| Synonyms | A2aR|ADORA2|RDC8 |
| Description | adenosine A2a receptor |
| Reference | MIM:102776|HGNC:HGNC:263|Ensembl:ENSG00000128271|Ensembl:ENSG00000258555|HPRD:00043|Vega:OTTHUMG00000150761 |
| Gene type | protein-coding |
| Map location | 22q11.23 |
| Pascal p-value | 0.434 |
| Sherlock p-value | 0.973 |
| Fetal beta | 0.578 |
| eGene | Myers' cis & trans |
| Support | GPCR SIGNALLING |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| GSMA_I | Genome scan meta-analysis | Psr: 0.031 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 4 | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.1128 |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs596985 | chr1 | 64277034 | ADORA2A | 135 | 0.15 | trans | ||
| rs16838771 | chr1 | 157511227 | ADORA2A | 135 | 2.199E-5 | trans | ||
| rs17575539 | chr2 | 62766126 | ADORA2A | 135 | 0.13 | trans | ||
| rs17019687 | chr2 | 80972176 | ADORA2A | 135 | 0.14 | trans | ||
| rs1881496 | chr2 | 233372354 | ADORA2A | 135 | 0.04 | trans | ||
| rs6803351 | chr3 | 17336986 | ADORA2A | 135 | 0.04 | trans | ||
| rs11916205 | chr3 | 17392363 | ADORA2A | 135 | 0.04 | trans | ||
| rs17043486 | chr3 | 17399005 | ADORA2A | 135 | 0.04 | trans | ||
| rs17029249 | chr3 | 17416384 | ADORA2A | 135 | 0.04 | trans | ||
| rs17043623 | chr3 | 17478890 | ADORA2A | 135 | 0.04 | trans | ||
| rs11128830 | chr3 | 17481727 | ADORA2A | 135 | 0.04 | trans | ||
| rs17044097 | chr3 | 17889750 | ADORA2A | 135 | 0.04 | trans | ||
| rs17044102 | chr3 | 17889838 | ADORA2A | 135 | 0.12 | trans | ||
| rs4627772 | 0 | ADORA2A | 135 | 0.06 | trans | |||
| rs17079580 | chr3 | 47363841 | ADORA2A | 135 | 0.06 | trans | ||
| rs12633404 | chr3 | 80381837 | ADORA2A | 135 | 0.01 | trans | ||
| rs12637719 | chr3 | 85328350 | ADORA2A | 135 | 2.888E-4 | trans | ||
| rs17647031 | chr3 | 116102744 | ADORA2A | 135 | 0.18 | trans | ||
| rs16864324 | chr3 | 189106780 | ADORA2A | 135 | 0.2 | trans | ||
| rs12651357 | chr4 | 53029658 | ADORA2A | 135 | 0.18 | trans | ||
| rs9996479 | chr4 | 53029771 | ADORA2A | 135 | 0.18 | trans | ||
| rs3025740 | chr4 | 114163593 | ADORA2A | 135 | 0.02 | trans | ||
| snp_a-4195294 | 0 | ADORA2A | 135 | 0.03 | trans | |||
| rs16894524 | chr5 | 65192799 | ADORA2A | 135 | 0.15 | trans | ||
| rs17459137 | chr5 | 114603454 | ADORA2A | 135 | 0.06 | trans | ||
| rs3812061 | chr5 | 126213763 | ADORA2A | 135 | 0.01 | trans | ||
| rs12655258 | chr5 | 126267350 | ADORA2A | 135 | 0.05 | trans | ||
| rs12652390 | chr5 | 126322101 | ADORA2A | 135 | 2.15E-4 | trans | ||
| rs17165249 | chr5 | 126395510 | ADORA2A | 135 | 0.12 | trans | ||
| rs17165234 | chr5 | 126403634 | ADORA2A | 135 | 3.703E-7 | trans | ||
| rs12655006 | chr5 | 126407601 | ADORA2A | 135 | 0.04 | trans | ||
| rs12656879 | chr5 | 126599075 | ADORA2A | 135 | 0.1 | trans | ||
| rs3733727 | chr5 | 134689299 | ADORA2A | 135 | 0.06 | trans | ||
| rs2270172 | chr6 | 30592413 | ADORA2A | 135 | 0.02 | trans | ||
| rs1041915 | chr6 | 130014252 | ADORA2A | 135 | 0.16 | trans | ||
| rs9397728 | chr6 | 150514248 | ADORA2A | 135 | 2.437E-4 | trans | ||
| rs11987753 | chr8 | 110166238 | ADORA2A | 135 | 0.04 | trans | ||
| rs10963890 | chr9 | 18977332 | ADORA2A | 135 | 0.07 | trans | ||
| rs6479279 | chr9 | 107476888 | ADORA2A | 135 | 0.16 | trans | ||
| rs2066315 | chr10 | 514137 | ADORA2A | 135 | 0.03 | trans | ||
| rs10794751 | chr10 | 1514218 | ADORA2A | 135 | 0.2 | trans | ||
| rs1440195 | chr11 | 116183393 | ADORA2A | 135 | 0.08 | trans | ||
| rs11178166 | chr12 | 70670585 | ADORA2A | 135 | 0.05 | trans | ||
| rs2297479 | chr12 | 125591249 | ADORA2A | 135 | 0.08 | trans | ||
| rs9603093 | chr13 | 37337849 | ADORA2A | 135 | 0.04 | trans | ||
| rs854406 | chr14 | 25204350 | ADORA2A | 135 | 0.11 | trans | ||
| rs8008199 | chr14 | 31002111 | ADORA2A | 135 | 0.1 | trans | ||
| rs4981760 | chr14 | 31002684 | ADORA2A | 135 | 0.02 | trans | ||
| rs230349 | chr14 | 31130389 | ADORA2A | 135 | 0.09 | trans | ||
| rs230365 | chr14 | 31139870 | ADORA2A | 135 | 0.16 | trans | ||
| rs11628947 | chr14 | 31161925 | ADORA2A | 135 | 0.16 | trans | ||
| rs17104965 | chr14 | 37145915 | ADORA2A | 135 | 0 | trans | ||
| rs17105036 | chr14 | 37154593 | ADORA2A | 135 | 0 | trans | ||
| rs362341 | chr14 | 73641457 | ADORA2A | 135 | 0.04 | trans | ||
| rs4564513 | chr15 | 57960009 | ADORA2A | 135 | 0.11 | trans | ||
| rs8056861 | chr16 | 56921039 | ADORA2A | 135 | 0 | trans | ||
| rs443222 | chr16 | 83889679 | ADORA2A | 135 | 0.01 | trans | ||
| rs824575 | chr16 | 83891459 | ADORA2A | 135 | 0.01 | trans | ||
| rs1056626 | chr17 | 4931552 | ADORA2A | 135 | 0.19 | trans | ||
| rs355306 | chr18 | 33105716 | ADORA2A | 135 | 0.01 | trans | ||
| rs7504799 | chr18 | 70057077 | ADORA2A | 135 | 0.19 | trans | ||
| rs10405894 | chr19 | 52631599 | ADORA2A | 135 | 0.12 | trans | ||
| rs6102879 | chr20 | 41138766 | ADORA2A | 135 | 0.18 | trans | ||
| rs2835891 | chr21 | 39056226 | ADORA2A | 135 | 0.01 | trans | ||
| rs1883665 | chrX | 17650382 | ADORA2A | 135 | 4.912E-6 | trans | ||
| rs7062071 | chrX | 17724830 | ADORA2A | 135 | 0.01 | trans | ||
| rs12690023 | chrX | 50830077 | ADORA2A | 135 | 0.15 | trans | ||
| rs7879274 | chrX | 145592957 | ADORA2A | 135 | 0.14 | trans |
Section II. Transcriptome annotation
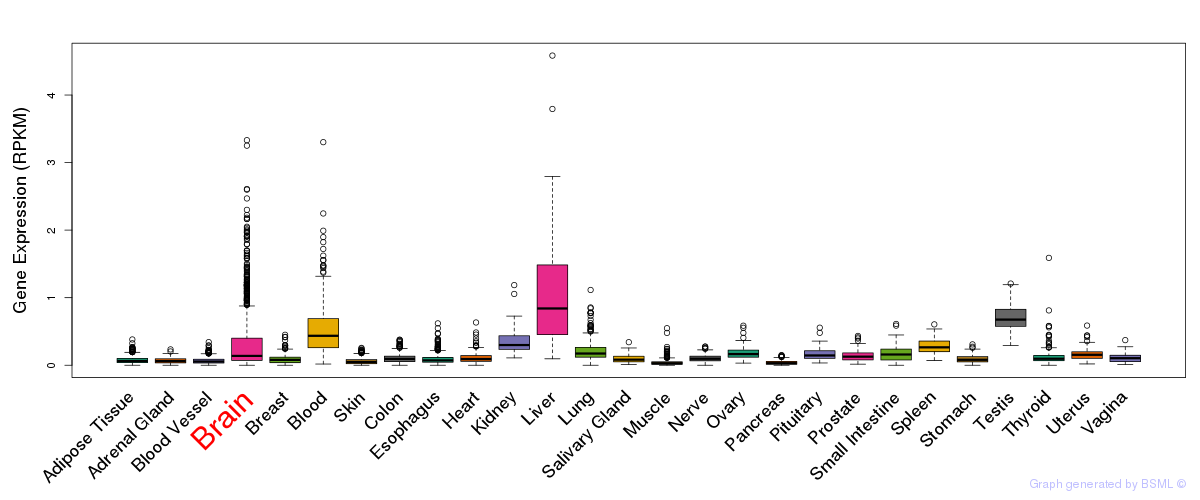
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0001609 | adenosine receptor activity, G-protein coupled | IEA | - | |
| GO:0004872 | receptor activity | IEA | - | |
| GO:0016519 | gastric inhibitory peptide receptor activity | NAS | 8522976 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007417 | central nervous system development | TAS | Brain (GO term level: 6) | 10945659 |
| GO:0032230 | positive regulation of synaptic transmission, GABAergic | IEA | neuron, GABA, Synap, Neurotransmitter (GO term level: 10) | - |
| GO:0001963 | synaptic transmission, dopaminergic | IEA | neuron, Synap, Neurotransmitter, dopamine (GO term level: 8) | - |
| GO:0006836 | neurotransmitter transport | NAS | neuron, Neurotransmitter (GO term level: 5) | 8522976 |
| GO:0001975 | response to amphetamine | IEA | - | |
| GO:0001973 | adenosine receptor signaling pathway | IEA | - | |
| GO:0007190 | activation of adenylate cyclase activity | EXP | 16805430 | |
| GO:0007189 | G-protein signaling, adenylate cyclase activating pathway | IEA | - | |
| GO:0007188 | G-protein signaling, coupled to cAMP nucleotide second messenger | TAS | 10899090 | |
| GO:0007186 | G-protein coupled receptor protein signaling pathway | IEA | - | |
| GO:0007169 | transmembrane receptor protein tyrosine kinase signaling pathway | EXP | 16939974 | |
| GO:0007267 | cell-cell signaling | TAS | 7818494 | |
| GO:0006171 | cAMP biosynthetic process | TAS | 10899090 | |
| GO:0006968 | cellular defense response | TAS | 10051547 | |
| GO:0007165 | signal transduction | IEA | - | |
| GO:0008015 | blood circulation | TAS | 9262401 | |
| GO:0007626 | locomotory behavior | IEA | - | |
| GO:0007600 | sensory perception | TAS | 9262401 | |
| GO:0007596 | blood coagulation | TAS | 10899090 | |
| GO:0006954 | inflammatory response | TAS | 8360491 | |
| GO:0006915 | apoptosis | TAS | 7818494 | |
| GO:0006909 | phagocytosis | TAS | 8360491 | |
| GO:0042755 | eating behavior | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005624 | membrane fraction | IEA | - | |
| GO:0005624 | membrane fraction | TAS | 10051547 | |
| GO:0016021 | integral to membrane | IEA | - | |
| GO:0005886 | plasma membrane | TAS | 8794889 |10051547 | |
| GO:0005887 | integral to plasma membrane | TAS | 8794889 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG CALCIUM SIGNALING PATHWAY | 178 | 134 | All SZGR 2.0 genes in this pathway |
| KEGG NEUROACTIVE LIGAND RECEPTOR INTERACTION | 272 | 195 | All SZGR 2.0 genes in this pathway |
| KEGG VASCULAR SMOOTH MUSCLE CONTRACTION | 115 | 81 | All SZGR 2.0 genes in this pathway |
| PID HIF2PATHWAY | 34 | 29 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALLING BY NGF | 217 | 167 | All SZGR 2.0 genes in this pathway |
| REACTOME NGF SIGNALLING VIA TRKA FROM THE PLASMA MEMBRANE | 137 | 105 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY GPCR | 920 | 449 | All SZGR 2.0 genes in this pathway |
| REACTOME CLASS A1 RHODOPSIN LIKE RECEPTORS | 305 | 177 | All SZGR 2.0 genes in this pathway |
| REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | 16 | 8 | All SZGR 2.0 genes in this pathway |
| REACTOME GPCR DOWNSTREAM SIGNALING | 805 | 368 | All SZGR 2.0 genes in this pathway |
| REACTOME G ALPHA S SIGNALLING EVENTS | 121 | 82 | All SZGR 2.0 genes in this pathway |
| REACTOME GPCR LIGAND BINDING | 408 | 246 | All SZGR 2.0 genes in this pathway |
| RASHI RESPONSE TO IONIZING RADIATION 5 | 147 | 89 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 2B | 392 | 251 | All SZGR 2.0 genes in this pathway |
| KOYAMA SEMA3B TARGETS DN | 411 | 249 | All SZGR 2.0 genes in this pathway |
| ROZANOV MMP14 TARGETS UP | 266 | 171 | All SZGR 2.0 genes in this pathway |
| FAELT B CLL WITH VH REARRANGEMENTS UP | 48 | 34 | All SZGR 2.0 genes in this pathway |
| MCCLUNG DELTA FOSB TARGETS 8WK | 47 | 38 | All SZGR 2.0 genes in this pathway |
| MCCLUNG COCAINE REWARD 5D | 79 | 62 | All SZGR 2.0 genes in this pathway |
| ZHANG ANTIVIRAL RESPONSE TO RIBAVIRIN DN | 51 | 32 | All SZGR 2.0 genes in this pathway |
| SAFFORD T LYMPHOCYTE ANERGY | 87 | 54 | All SZGR 2.0 genes in this pathway |
| MAHAJAN RESPONSE TO IL1A UP | 81 | 52 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 STIMULATED | 1022 | 619 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 UNSTIMULATED | 1229 | 713 | All SZGR 2.0 genes in this pathway |
| FOSTER TOLERANT MACROPHAGE UP | 156 | 92 | All SZGR 2.0 genes in this pathway |
| YAUCH HEDGEHOG SIGNALING PARACRINE DN | 264 | 159 | All SZGR 2.0 genes in this pathway |
| LINDSTEDT DENDRITIC CELL MATURATION B | 53 | 36 | All SZGR 2.0 genes in this pathway |
| MEISSNER NPC HCP WITH H3 UNMETHYLATED | 536 | 296 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K4ME3 AND H3K27ME3 | 1069 | 729 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MCV6 HCP WITH H3K27ME3 | 435 | 318 | All SZGR 2.0 genes in this pathway |
| WIERENGA STAT5A TARGETS DN | 213 | 127 | All SZGR 2.0 genes in this pathway |
| CERIBELLI GENES INACTIVE AND BOUND BY NFY | 45 | 27 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 2ND EGF PULSE ONLY | 1725 | 838 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-214 | 537 | 543 | m8 | hsa-miR-214brain | ACAGCAGGCACAGACAGGCAG |
| miR-34b | 786 | 792 | m8 | hsa-miR-34b | UAGGCAGUGUCAUUAGCUGAUUG |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.