Gene Page: COX10
Summary ?
| GeneID | 1352 |
| Symbol | COX10 |
| Synonyms | - |
| Description | COX10 heme A:farnesyltransferase cytochrome c oxidase assembly factor |
| Reference | MIM:602125|HGNC:HGNC:2260|Ensembl:ENSG00000006695|HPRD:03673|Vega:OTTHUMG00000058814 |
| Gene type | protein-coding |
| Map location | 17p12 |
| Pascal p-value | 0.092 |
| Sherlock p-value | 0.836 |
| Fetal beta | 0.149 |
| DMG | 2 (# studies) |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 2 |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 2 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg27005118 | 17 | 13972210 | COX10 | 2.861E-4 | 0.484 | 0.039 | DMG:Wockner_2014 |
| cg22956598 | 17 | 13973019 | COX10 | 2.27E-9 | -0.02 | 1.74E-6 | DMG:Jaffe_2016 |
Section II. Transcriptome annotation
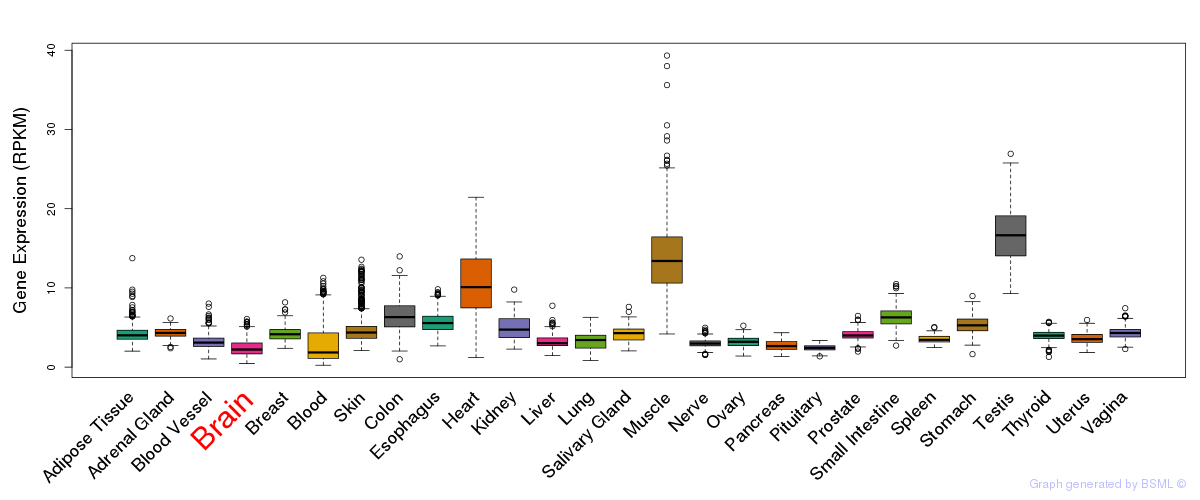
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| TARBP1 | 0.45 | 0.38 |
| PDIA2 | 0.43 | 0.31 |
| PLEKHH2 | 0.43 | 0.28 |
| TAS2R49 | 0.43 | 0.28 |
| SEC31B | 0.43 | 0.34 |
| AC022382.1 | 0.42 | 0.32 |
| MYO15B | 0.42 | 0.32 |
| PLA2G4B | 0.42 | 0.29 |
| AC008443.1 | 0.42 | 0.25 |
| AC005841.1 | 0.41 | 0.22 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| GAPDH | -0.24 | -0.21 |
| SLC3A2 | -0.24 | -0.23 |
| FBLN2 | -0.22 | -0.19 |
| BTBD17 | -0.22 | -0.20 |
| RASIP1 | -0.22 | -0.20 |
| HLA-A | -0.22 | -0.25 |
| ESAM | -0.21 | -0.17 |
| CHCHD9 | -0.21 | -0.22 |
| S1PR4 | -0.21 | -0.20 |
| BSG | -0.21 | -0.21 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG OXIDATIVE PHOSPHORYLATION | 135 | 73 | All SZGR 2.0 genes in this pathway |
| KEGG PORPHYRIN AND CHLOROPHYLL METABOLISM | 41 | 23 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN DN | 584 | 395 | All SZGR 2.0 genes in this pathway |
| AMIT SERUM RESPONSE 480 MCF10A | 39 | 20 | All SZGR 2.0 genes in this pathway |
| MOOTHA VOXPHOS | 87 | 51 | All SZGR 2.0 genes in this pathway |
| SANSOM APC MYC TARGETS | 217 | 138 | All SZGR 2.0 genes in this pathway |
| STEIN ESRRA TARGETS UP | 388 | 234 | All SZGR 2.0 genes in this pathway |
| GRADE COLON CANCER UP | 871 | 505 | All SZGR 2.0 genes in this pathway |
| BLUM RESPONSE TO SALIRASIB UP | 245 | 159 | All SZGR 2.0 genes in this pathway |
| MOOTHA HUMAN MITODB 6 2002 | 429 | 260 | All SZGR 2.0 genes in this pathway |
| MOOTHA MITOCHONDRIA | 447 | 277 | All SZGR 2.0 genes in this pathway |
| HSIAO HOUSEKEEPING GENES | 389 | 245 | All SZGR 2.0 genes in this pathway |
| STEIN ESRRA TARGETS | 535 | 325 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| LINSLEY MIR16 TARGETS | 206 | 127 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS DN | 882 | 538 | All SZGR 2.0 genes in this pathway |
| KIM TIAL1 TARGETS | 32 | 22 | All SZGR 2.0 genes in this pathway |