Gene Page: CPE
Summary ?
| GeneID | 1363 |
| Symbol | CPE |
| Synonyms | CPH |
| Description | carboxypeptidase E |
| Reference | MIM:114855|HGNC:HGNC:2303|Ensembl:ENSG00000109472|HPRD:00272|Vega:OTTHUMG00000150252 |
| Gene type | protein-coding |
| Map location | 4q32.3 |
| Pascal p-value | 0.052 |
| Sherlock p-value | 0.002 |
| Fetal beta | -0.937 |
| Support | CompositeSet Darnell FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenics,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
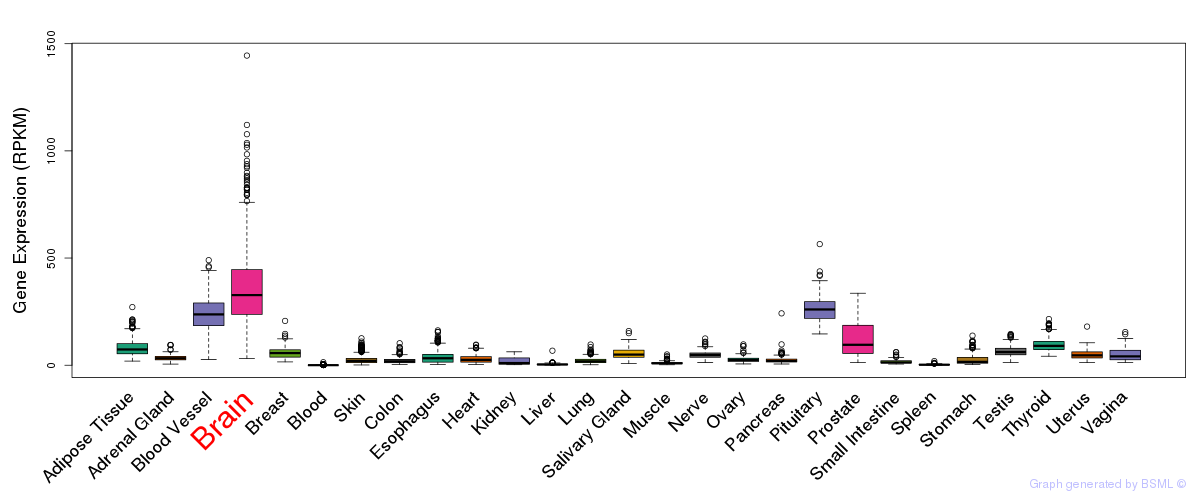
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0004181 | metallocarboxypeptidase activity | IEA | - | |
| GO:0008270 | zinc ion binding | IEA | - | |
| GO:0008237 | metallopeptidase activity | IEA | - | |
| GO:0008233 | peptidase activity | IEA | - | |
| GO:0046872 | metal ion binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007218 | neuropeptide signaling pathway | NAS | Neurotransmitter (GO term level: 8) | 2334405 |
| GO:0006464 | protein modification process | NAS | 2334405 | |
| GO:0006508 | proteolysis | IEA | - | |
| GO:0008152 | metabolic process | TAS | 7663508 | |
| GO:0030070 | insulin processing | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005576 | extracellular region | IEA | - | |
| GO:0005886 | plasma membrane | TAS | 9019408 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| C1orf103 | FLJ11269 | RIF1 | RP11-96K19.1 | chromosome 1 open reading frame 103 | Two-hybrid | BioGRID | 16169070 |
| CHD3 | Mi-2a | Mi2-ALPHA | ZFH | chromodomain helicase DNA binding protein 3 | Two-hybrid | BioGRID | 16169070 |
| GCG | GLP1 | GLP2 | GRPP | glucagon | - | HPRD | 11375130 |
| GTF3C1 | DKFZp686A111 | TFIIIC | TFIIIC220 | TFIIICalpha | general transcription factor IIIC, polypeptide 1, alpha 220kDa | Two-hybrid | BioGRID | 16169070 |
| MED31 | 3110004H13Rik | CGI-125 | FLJ27436 | FLJ36714 | Soh1 | mediator complex subunit 31 | Two-hybrid | BioGRID | 16169070 |
| PMCH | MCH | pro-melanin-concentrating hormone | - | HPRD | 8770919 |
| POLA2 | FLJ21662 | FLJ37250 | polymerase (DNA directed), alpha 2 (70kD subunit) | Two-hybrid | BioGRID | 16169070 |
| POLDIP2 | DKFZp586F1524 | PDIP38 | POLD4 | polymerase (DNA-directed), delta interacting protein 2 | Two-hybrid | BioGRID | 16169070 |
| ROBO2 | KIAA1568 | SAX3 | roundabout, axon guidance receptor, homolog 2 (Drosophila) | Two-hybrid | BioGRID | 16169070 |
| RPA1 | HSSB | REPA1 | RF-A | RP-A | RPA70 | replication protein A1, 70kDa | Two-hybrid | BioGRID | 16169070 |
| UTP14A | KIAA0266 | NY-CO-16 | SDCCAG16 | dJ537K23.3 | UTP14, U3 small nucleolar ribonucleoprotein, homolog A (yeast) | Two-hybrid | BioGRID | 16169070 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG TYPE I DIABETES MELLITUS | 44 | 38 | All SZGR 2.0 genes in this pathway |
| PID ARF6 TRAFFICKING PATHWAY | 49 | 34 | All SZGR 2.0 genes in this pathway |
| REACTOME DIABETES PATHWAYS | 133 | 91 | All SZGR 2.0 genes in this pathway |
| REACTOME INSULIN SYNTHESIS AND PROCESSING | 21 | 15 | All SZGR 2.0 genes in this pathway |
| NAKAMURA TUMOR ZONE PERIPHERAL VS CENTRAL DN | 634 | 384 | All SZGR 2.0 genes in this pathway |
| WANG CLIM2 TARGETS DN | 186 | 114 | All SZGR 2.0 genes in this pathway |
| VECCHI GASTRIC CANCER ADVANCED VS EARLY UP | 175 | 120 | All SZGR 2.0 genes in this pathway |
| VECCHI GASTRIC CANCER EARLY DN | 367 | 220 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC1 TARGETS DN | 260 | 143 | All SZGR 2.0 genes in this pathway |
| WAMUNYOKOLI OVARIAN CANCER LMP DN | 199 | 124 | All SZGR 2.0 genes in this pathway |
| WAMUNYOKOLI OVARIAN CANCER GRADES 1 2 DN | 67 | 43 | All SZGR 2.0 genes in this pathway |
| MAYBURD RESPONSE TO L663536 DN | 56 | 32 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA POORLY DIFFERENTIATED DN | 805 | 505 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA ANAPLASTIC DN | 537 | 339 | All SZGR 2.0 genes in this pathway |
| GOZGIT ESR1 TARGETS DN | 781 | 465 | All SZGR 2.0 genes in this pathway |
| CHEBOTAEV GR TARGETS DN | 120 | 73 | All SZGR 2.0 genes in this pathway |
| PEREZ TP53 TARGETS | 1174 | 695 | All SZGR 2.0 genes in this pathway |
| KANG GIST WITH PDGFRA UP | 50 | 27 | All SZGR 2.0 genes in this pathway |
| BORLAK LIVER CANCER EGF UP | 57 | 41 | All SZGR 2.0 genes in this pathway |
| MOHANKUMAR TLX1 TARGETS UP | 414 | 287 | All SZGR 2.0 genes in this pathway |
| GROSS ELK3 TARGETS UP | 27 | 16 | All SZGR 2.0 genes in this pathway |
| COLIN PILOCYTIC ASTROCYTOMA VS GLIOBLASTOMA UP | 35 | 32 | All SZGR 2.0 genes in this pathway |
| GOTZMANN EPITHELIAL TO MESENCHYMAL TRANSITION DN | 206 | 136 | All SZGR 2.0 genes in this pathway |
| ROZANOV MMP14 TARGETS UP | 266 | 171 | All SZGR 2.0 genes in this pathway |
| BECKER TAMOXIFEN RESISTANCE UP | 50 | 36 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER DENA UP | 60 | 40 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER E2F1 UP | 62 | 35 | All SZGR 2.0 genes in this pathway |
| BRUNO HEMATOPOIESIS | 66 | 48 | All SZGR 2.0 genes in this pathway |
| JIANG AGING CEREBRAL CORTEX UP | 36 | 27 | All SZGR 2.0 genes in this pathway |
| KAAB HEART ATRIUM VS VENTRICLE UP | 249 | 170 | All SZGR 2.0 genes in this pathway |
| JIANG AGING HYPOTHALAMUS DN | 40 | 31 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 48HR UP | 180 | 125 | All SZGR 2.0 genes in this pathway |
| GAJATE RESPONSE TO TRABECTEDIN UP | 67 | 45 | All SZGR 2.0 genes in this pathway |
| WEIGEL OXIDATIVE STRESS BY HNE AND TBH | 60 | 42 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS 7 | 51 | 34 | All SZGR 2.0 genes in this pathway |
| KEEN RESPONSE TO ROSIGLITAZONE DN | 106 | 68 | All SZGR 2.0 genes in this pathway |
| XU GH1 AUTOCRINE TARGETS UP | 268 | 157 | All SZGR 2.0 genes in this pathway |
| WANG SMARCE1 TARGETS UP | 280 | 183 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 4 | 307 | 185 | All SZGR 2.0 genes in this pathway |
| LEIN LOCALIZED TO DISTAL AND PROXIMAL DENDRITES | 17 | 12 | All SZGR 2.0 genes in this pathway |
| GAVIN FOXP3 TARGETS CLUSTER P2 | 79 | 55 | All SZGR 2.0 genes in this pathway |
| ZHENG BOUND BY FOXP3 | 491 | 310 | All SZGR 2.0 genes in this pathway |
| ZHENG FOXP3 TARGETS IN THYMUS UP | 196 | 137 | All SZGR 2.0 genes in this pathway |
| ZHENG FOXP3 TARGETS IN T LYMPHOCYTE DN | 37 | 29 | All SZGR 2.0 genes in this pathway |
| WANG LSD1 TARGETS DN | 39 | 30 | All SZGR 2.0 genes in this pathway |
| KONDO EZH2 TARGETS | 245 | 148 | All SZGR 2.0 genes in this pathway |
| MASSARWEH TAMOXIFEN RESISTANCE DN | 258 | 160 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER UP | 973 | 570 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER NORMAL LIKE UP | 476 | 285 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL DN | 701 | 446 | All SZGR 2.0 genes in this pathway |
| MATZUK CENTRAL FOR FEMALE FERTILITY | 29 | 25 | All SZGR 2.0 genes in this pathway |
| BOQUEST STEM CELL CULTURED VS FRESH UP | 425 | 298 | All SZGR 2.0 genes in this pathway |
| LABBE WNT3A TARGETS UP | 112 | 71 | All SZGR 2.0 genes in this pathway |
| LABBE TGFB1 TARGETS UP | 102 | 64 | All SZGR 2.0 genes in this pathway |
| LABBE TARGETS OF TGFB1 AND WNT3A UP | 111 | 70 | All SZGR 2.0 genes in this pathway |
| BOYLAN MULTIPLE MYELOMA C D DN | 252 | 155 | All SZGR 2.0 genes in this pathway |
| WU SILENCED BY METHYLATION IN BLADDER CANCER | 55 | 42 | All SZGR 2.0 genes in this pathway |
| HELLEBREKERS SILENCED DURING TUMOR ANGIOGENESIS | 80 | 56 | All SZGR 2.0 genes in this pathway |
| SWEET LUNG CANCER KRAS DN | 435 | 289 | All SZGR 2.0 genes in this pathway |
| KOBAYASHI EGFR SIGNALING 24HR UP | 101 | 65 | All SZGR 2.0 genes in this pathway |
| BOYLAN MULTIPLE MYELOMA PCA1 UP | 101 | 66 | All SZGR 2.0 genes in this pathway |
| KIM BIPOLAR DISORDER OLIGODENDROCYTE DENSITY CORR UP | 682 | 440 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS CONFLUENT | 567 | 365 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 2 DN | 336 | 211 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP D | 280 | 158 | All SZGR 2.0 genes in this pathway |
| PASINI SUZ12 TARGETS DN | 315 | 215 | All SZGR 2.0 genes in this pathway |
| AZARE NEOPLASTIC TRANSFORMATION BY STAT3 DN | 17 | 10 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS UP | 745 | 475 | All SZGR 2.0 genes in this pathway |
| FEVR CTNNB1 TARGETS UP | 682 | 433 | All SZGR 2.0 genes in this pathway |
| PEDERSEN METASTASIS BY ERBB2 ISOFORM 4 | 110 | 66 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG BOUND 8D | 658 | 397 | All SZGR 2.0 genes in this pathway |
| FOSTER KDM1A TARGETS DN | 211 | 119 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 1ST EGF PULSE ONLY | 1839 | 928 | All SZGR 2.0 genes in this pathway |