Gene Page: CRMP1
Summary ?
| GeneID | 1400 |
| Symbol | CRMP1 |
| Synonyms | CRMP-1|DPYSL1|DRP-1|DRP1|ULIP-3 |
| Description | collapsin response mediator protein 1 |
| Reference | MIM:602462|HGNC:HGNC:2365|Ensembl:ENSG00000072832|HPRD:03913|Vega:OTTHUMG00000125489 |
| Gene type | protein-coding |
| Map location | 4p16.1 |
| Pascal p-value | 0.27 |
| Sherlock p-value | 0.98 |
| Fetal beta | 1.313 |
| Support | CELL ADHESION AND TRANSSYNAPTIC SIGNALING G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-CONSENSUS G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL G2Cdb.human_BAYES-COLLINS-MOUSE-PSD-CONSENSUS G2Cdb.human_clathrin G2Cdb.human_Synaptosome G2Cdb.humanPSD G2Cdb.humanPSP CompositeSet Darnell FMRP targets Potential synaptic genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 3 | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0069 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
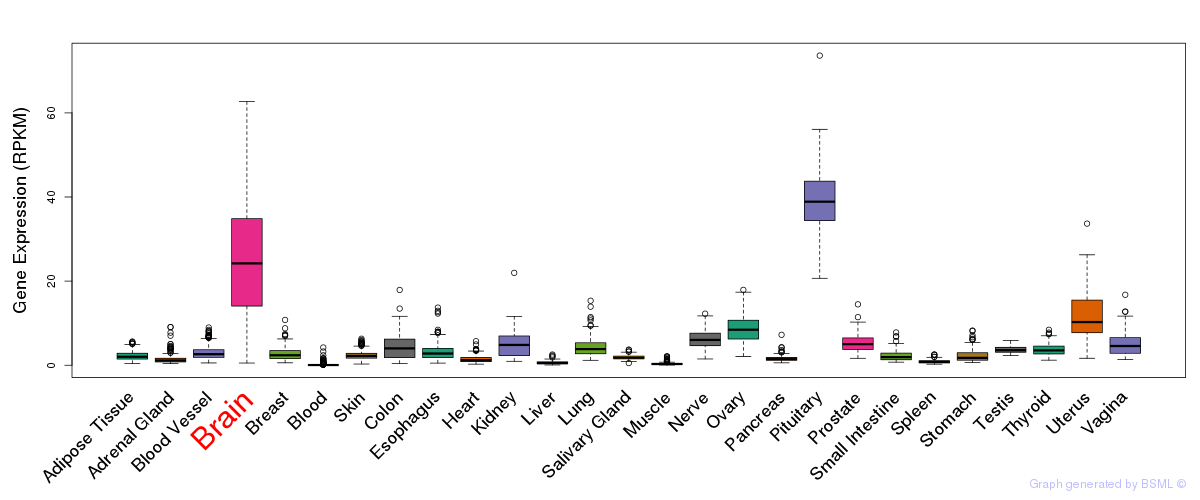
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0004157 | dihydropyrimidinase activity | TAS | 8973361 | |
| GO:0005515 | protein binding | IPI | 15383276 |16169070 | |
| GO:0016810 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007399 | nervous system development | TAS | neurite (GO term level: 5) | 8973361 |
| GO:0006139 | nucleobase, nucleoside, nucleotide and nucleic acid metabolic process | TAS | 8973361 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0043025 | cell soma | IEA | axon, dendrite (GO term level: 4) | - |
| GO:0030425 | dendrite | IEA | neuron, axon, dendrite (GO term level: 6) | - |
| GO:0005737 | cytoplasm | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| AGR2 | AG2 | GOB-4 | HAG-2 | XAG-2 | anterior gradient homolog 2 (Xenopus laevis) | Two-hybrid | BioGRID | 16169070 |
| ALDH2 | ALDH-E2 | ALDHI | ALDM | MGC1806 | aldehyde dehydrogenase 2 family (mitochondrial) | Two-hybrid | BioGRID | 16169070 |
| ARL15 | ARFRP2 | FLJ20051 | ADP-ribosylation factor-like 15 | Two-hybrid | BioGRID | 16169070 |
| AXIN1 | AXIN | MGC52315 | axin 1 | Two-hybrid | BioGRID | 16169070 |
| BTBD2 | - | BTB (POZ) domain containing 2 | Two-hybrid | BioGRID | 16169070 |
| CCDC106 | HSU79303 | ZNF581 | coiled-coil domain containing 106 | Two-hybrid | BioGRID | 16169070 |
| CCL18 | AMAC-1 | AMAC1 | CKb7 | DC-CK1 | DCCK1 | MIP-4 | PARC | SCYA18 | chemokine (C-C motif) ligand 18 (pulmonary and activation-regulated) | Two-hybrid | BioGRID | 16169070 |
| CCT7 | CCT-ETA | Ccth | MGC110985 | Nip7-1 | TCP-1-eta | chaperonin containing TCP1, subunit 7 (eta) | Two-hybrid | BioGRID | 16169070 |
| CDK5RAP2 | C48 | Cep215 | DKFZp686B1070 | DKFZp686D1070 | KIAA1633 | MCPH3 | CDK5 regulatory subunit associated protein 2 | Two-hybrid | BioGRID | 16169070 |
| CDK5RAP3 | C53 | HSF-27 | IC53 | LZAP | MST016 | OK/SW-cl.114 | CDK5 regulatory subunit associated protein 3 | Two-hybrid | BioGRID | 16169070 |
| DPYSL2 | CRMP2 | DHPRP2 | DRP-2 | DRP2 | dihydropyrimidinase-like 2 | - | HPRD,BioGRID | 12482610 |
| EEF1D | EF-1D | EF1D | FLJ20897 | FP1047 | eukaryotic translation elongation factor 1 delta (guanine nucleotide exchange protein) | Two-hybrid | BioGRID | 16169070 |
| EIF2S2 | DKFZp686L18198 | EIF2 | EIF2B | EIF2beta | MGC8508 | eukaryotic translation initiation factor 2, subunit 2 beta, 38kDa | Two-hybrid | BioGRID | 16169070 |
| EPN1 | - | epsin 1 | Two-hybrid | BioGRID | 16169070 |
| FAS | ALPS1A | APO-1 | APT1 | CD95 | FAS1 | FASTM | TNFRSF6 | Fas (TNF receptor superfamily, member 6) | Two-hybrid | BioGRID | 16169070 |
| FTH1 | FHC | FTH | FTHL6 | MGC104426 | PIG15 | PLIF | ferritin, heavy polypeptide 1 | Two-hybrid | BioGRID | 16169070 |
| FUBP1 | FBP | FUBP | far upstream element (FUSE) binding protein 1 | Two-hybrid | BioGRID | 16169070 |
| FXR1 | - | fragile X mental retardation, autosomal homolog 1 | Two-hybrid | BioGRID | 16169070 |
| HMGB1 | DKFZp686A04236 | HMG1 | HMG3 | SBP-1 | high-mobility group box 1 | Two-hybrid | BioGRID | 16169070 |
| HNRNPH1 | DKFZp686A15170 | HNRPH | HNRPH1 | hnRNPH | heterogeneous nuclear ribonucleoprotein H1 (H) | Two-hybrid | BioGRID | 16169070 |
| HNRNPH3 | 2H9 | FLJ34092 | HNRPH3 | heterogeneous nuclear ribonucleoprotein H3 (2H9) | Two-hybrid | BioGRID | 16169070 |
| HNRNPUL1 | E1B-AP5 | E1BAP5 | FLJ12944 | HNRPUL1 | heterogeneous nuclear ribonucleoprotein U-like 1 | Two-hybrid | BioGRID | 16169070 |
| HSPE1 | CPN10 | GROES | HSP10 | heat shock 10kDa protein 1 (chaperonin 10) | Two-hybrid | BioGRID | 16169070 |
| KLHL20 | KHLHX | KLEIP | KLHLX | RP3-383J4.3 | kelch-like 20 (Drosophila) | Two-hybrid | BioGRID | 16169070 |
| LRRC1 | FLJ10775 | FLJ11834 | LANO | dJ523E19.1 | leucine rich repeat containing 1 | Two-hybrid | BioGRID | 16169070 |
| LSM2 | C6orf28 | G7b | YBL026W | snRNP | LSM2 homolog, U6 small nuclear RNA associated (S. cerevisiae) | Two-hybrid | BioGRID | 16169070 |
| MOBKL3 | 2C4D | CGI-95 | MGC12264 | MOB1 | MOB3 | PREI3 | MOB1, Mps One Binder kinase activator-like 3 (yeast) | Two-hybrid | BioGRID | 16169070 |
| MRPS12 | MPR-S12 | MT-RPS12 | RPMS12 | RPS12 | RPSM12 | mitochondrial ribosomal protein S12 | Two-hybrid | BioGRID | 16169070 |
| NAT9 | DKFZp564C103 | EBSP | N-acetyltransferase 9 (GCN5-related, putative) | Two-hybrid | BioGRID | 16169070 |
| NDUFV2 | - | NADH dehydrogenase (ubiquinone) flavoprotein 2, 24kDa | Two-hybrid | BioGRID | 16169070 |
| NVL | - | nuclear VCP-like | Two-hybrid | BioGRID | 16169070 |
| PAFAH1B3 | - | platelet-activating factor acetylhydrolase, isoform Ib, gamma subunit 29kDa | Two-hybrid | BioGRID | 16169070 |
| PFN1 | - | profilin 1 | Two-hybrid | BioGRID | 16169070 |
| PMF1 | - | polyamine-modulated factor 1 | Two-hybrid | BioGRID | 16169070 |
| PPP1R8 | ARD-1 | ARD1 | NIPP-1 | NIPP1 | PRO2047 | protein phosphatase 1, regulatory (inhibitor) subunit 8 | Two-hybrid | BioGRID | 16169070 |
| PSMD11 | MGC3844 | Rpn6 | S9 | p44.5 | proteasome (prosome, macropain) 26S subunit, non-ATPase, 11 | Two-hybrid | BioGRID | 16169070 |
| RGL2 | HKE1.5 | KE1.5 | RAB2L | ral guanine nucleotide dissociation stimulator-like 2 | Two-hybrid | BioGRID | 16169070 |
| RHOA | ARH12 | ARHA | RHO12 | RHOH12 | ras homolog gene family, member A | Phenotypic Suppression | BioGRID | 12482610 |
| RPA2 | REPA2 | RPA32 | replication protein A2, 32kDa | Two-hybrid | BioGRID | 16169070 |
| RTN4 | ASY | NI220/250 | NOGO | NOGO-A | NOGOC | NSP | NSP-CL | Nbla00271 | Nbla10545 | Nogo-B | Nogo-C | RTN-X | RTN4-A | RTN4-B1 | RTN4-B2 | RTN4-C | reticulon 4 | Two-hybrid | BioGRID | 16169070 |
| SAT1 | DC21 | KFSD | SAT | SSAT | SSAT-1 | spermidine/spermine N1-acetyltransferase 1 | Two-hybrid | BioGRID | 16169070 |
| SEPHS1 | MGC4980 | SELD | SPS | SPS1 | selenophosphate synthetase 1 | Two-hybrid | BioGRID | 16169070 |
| SERPINB9 | CAP-3 | CAP3 | PI9 | serpin peptidase inhibitor, clade B (ovalbumin), member 9 | Two-hybrid | BioGRID | 16169070 |
| SNRPG | MGC117317 | SMG | small nuclear ribonucleoprotein polypeptide G | Two-hybrid | BioGRID | 16169070 |
| TFG | FLJ36137 | TF6 | TRKT3 | TRK-fused gene | Two-hybrid | BioGRID | 16169070 |
| TK1 | TK2 | thymidine kinase 1, soluble | Two-hybrid | BioGRID | 16169070 |
| TRIP13 | 16E1BP | thyroid hormone receptor interactor 13 | Two-hybrid | BioGRID | 16169070 |
| UBE2B | E2-17kDa | HHR6B | HR6B | RAD6B | UBC2 | ubiquitin-conjugating enzyme E2B (RAD6 homolog) | Two-hybrid | BioGRID | 16169070 |
| ZNF24 | KOX17 | RSG-A | ZNF191 | ZSCAN3 | Zfp191 | zinc finger protein 24 | Two-hybrid | BioGRID | 16169070 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| REACTOME DEVELOPMENTAL BIOLOGY | 396 | 292 | All SZGR 2.0 genes in this pathway |
| REACTOME AXON GUIDANCE | 251 | 188 | All SZGR 2.0 genes in this pathway |
| REACTOME CRMPS IN SEMA3A SIGNALING | 14 | 14 | All SZGR 2.0 genes in this pathway |
| REACTOME SEMAPHORIN INTERACTIONS | 68 | 53 | All SZGR 2.0 genes in this pathway |
| DAVICIONI MOLECULAR ARMS VS ERMS UP | 332 | 228 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC1 TARGETS DN | 260 | 143 | All SZGR 2.0 genes in this pathway |
| VANHARANTA UTERINE FIBROID UP | 45 | 26 | All SZGR 2.0 genes in this pathway |
| GAUSSMANN MLL AF4 FUSION TARGETS G UP | 238 | 135 | All SZGR 2.0 genes in this pathway |
| LASTOWSKA NEUROBLASTOMA COPY NUMBER DN | 800 | 473 | All SZGR 2.0 genes in this pathway |
| GUENTHER GROWTH SPHERICAL VS ADHERENT UP | 21 | 15 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 12HR DN | 57 | 45 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER SOX9 TARGETS IN PROSTATE DEVELOPMENT DN | 45 | 33 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES 1 | 379 | 235 | All SZGR 2.0 genes in this pathway |
| BENPORATH SUZ12 TARGETS | 1038 | 678 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES WITH H3K27ME3 | 1118 | 744 | All SZGR 2.0 genes in this pathway |
| POMEROY MEDULLOBLASTOMA PROGNOSIS UP | 47 | 30 | All SZGR 2.0 genes in this pathway |
| PETROVA ENDOTHELIUM LYMPHATIC VS BLOOD UP | 131 | 87 | All SZGR 2.0 genes in this pathway |
| RORIE TARGETS OF EWSR1 FLI1 FUSION DN | 30 | 23 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 48HR DN | 504 | 323 | All SZGR 2.0 genes in this pathway |
| DOUGLAS BMI1 TARGETS UP | 566 | 371 | All SZGR 2.0 genes in this pathway |
| AMUNDSON GAMMA RADIATION RESISTANCE | 20 | 14 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER WITH H3K27ME3 DN | 228 | 114 | All SZGR 2.0 genes in this pathway |
| MOOTHA PGC | 420 | 269 | All SZGR 2.0 genes in this pathway |
| GOLDRATH ANTIGEN RESPONSE | 346 | 192 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS OLIGODENDROCYTE NUMBER CORR UP | 756 | 494 | All SZGR 2.0 genes in this pathway |
| KIM BIPOLAR DISORDER OLIGODENDROCYTE DENSITY CORR UP | 682 | 440 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS CALB1 CORR UP | 548 | 370 | All SZGR 2.0 genes in this pathway |
| VERHAAK GLIOBLASTOMA PRONEURAL | 177 | 132 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP D | 280 | 158 | All SZGR 2.0 genes in this pathway |
| PASINI SUZ12 TARGETS DN | 315 | 215 | All SZGR 2.0 genes in this pathway |
| HOLLEMAN VINCRISTINE RESISTANCE B ALL UP | 38 | 22 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-329 | 167 | 173 | m8 | hsa-miR-329brain | AACACACCUGGUUAACCUCUUU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.